[English] 日本語
 Yorodumi
Yorodumi- EMDB-3293: Sub-tomogram averaging of Lassa virus glycoprotein spike from vir... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3293 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Sub-tomogram averaging of Lassa virus glycoprotein spike from virus-like particles at pH 5 in complex with purified LAMP1 fragment | |||||||||
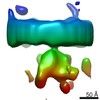
 Map data Map data | Sub-tomogram average of the glycoprotein spike trimer-receptor fragment complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | lassa virus / membrane protein / glycoprotein / receptor binding / membrane fusion / lysosome-associated membrane protein 1 / LAMP1 / receptor / complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of organelle transport along microtubule / positive regulation of natural killer cell degranulation / granzyme-mediated programmed cell death signaling pathway / phagolysosome membrane / cytolytic granule membrane / Golgi to lysosome transport / establishment of protein localization to organelle / lysosomal lumen acidification / positive regulation of natural killer cell mediated cytotoxicity / azurophil granule membrane ...regulation of organelle transport along microtubule / positive regulation of natural killer cell degranulation / granzyme-mediated programmed cell death signaling pathway / phagolysosome membrane / cytolytic granule membrane / Golgi to lysosome transport / establishment of protein localization to organelle / lysosomal lumen acidification / positive regulation of natural killer cell mediated cytotoxicity / azurophil granule membrane / ion channel inhibitor activity / autolysosome / autophagosome membrane / ficolin-1-rich granule membrane / multivesicular body / sarcolemma / melanosome / late endosome / synaptic vesicle / late endosome membrane / virus receptor activity / lysosome / protein stabilization / endosome membrane / protein domain specific binding / lysosomal membrane / external side of plasma membrane / Neutrophil degranulation / perinuclear region of cytoplasm / enzyme binding / extracellular exosome / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Lassa virus Lassa virus | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 14.8 Å | |||||||||
 Authors Authors | Li S / Zhaoyang S / Pryce R / Parsy M-L / Fehling SK / Schlie K / Siebert CA / Garten W / Bowden TA / Strecker T / Huiskonen JT | |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2016 Journal: PLoS Pathog / Year: 2016Title: Acidic pH-Induced Conformations and LAMP1 Binding of the Lassa Virus Glycoprotein Spike. Authors: Sai Li / Zhaoyang Sun / Rhys Pryce / Marie-Laure Parsy / Sarah K Fehling / Katrin Schlie / C Alistair Siebert / Wolfgang Garten / Thomas A Bowden / Thomas Strecker / Juha T Huiskonen /   Abstract: Lassa virus is an enveloped, bi-segmented RNA virus and the most prevalent and fatal of all Old World arenaviruses. Virus entry into the host cell is mediated by a tripartite surface spike complex, ...Lassa virus is an enveloped, bi-segmented RNA virus and the most prevalent and fatal of all Old World arenaviruses. Virus entry into the host cell is mediated by a tripartite surface spike complex, which is composed of two viral glycoprotein subunits, GP1 and GP2, and the stable signal peptide. Of these, GP1 binds to cellular receptors and GP2 catalyzes fusion between the viral envelope and the host cell membrane during endocytosis. The molecular structure of the spike and conformational rearrangements induced by low pH, prior to fusion, remain poorly understood. Here, we analyzed the three-dimensional ultrastructure of Lassa virus using electron cryotomography. Sub-tomogram averaging yielded a structure of the glycoprotein spike at 14-Å resolution. The spikes are trimeric, cover the virion envelope, and connect to the underlying matrix. Structural changes to the spike, following acidification, support a viral entry mechanism dependent on binding to the lysosome-resident receptor LAMP1 and further dissociation of the membrane-distal GP1 subunits. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3293.map.gz emd_3293.map.gz | 7.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3293-v30.xml emd-3293-v30.xml emd-3293.xml emd-3293.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3293_fsc.xml emd_3293_fsc.xml | 4.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_3293.tif emd_3293.tif | 325.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3293 http://ftp.pdbj.org/pub/emdb/structures/EMD-3293 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3293 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3293 | HTTPS FTP |
-Validation report
| Summary document |  emd_3293_validation.pdf.gz emd_3293_validation.pdf.gz | 247.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3293_full_validation.pdf.gz emd_3293_full_validation.pdf.gz | 246.7 KB | Display | |
| Data in XML |  emd_3293_validation.xml.gz emd_3293_validation.xml.gz | 8.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3293 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3293 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3293 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3293 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3293.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3293.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sub-tomogram average of the glycoprotein spike trimer-receptor fragment complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Purified Lassa virus VLPs at pH 5 mixed with purified fragment of...
| Entire | Name: Purified Lassa virus VLPs at pH 5 mixed with purified fragment of LAMP1 |
|---|---|
| Components |
|
-Supramolecule #1000: Purified Lassa virus VLPs at pH 5 mixed with purified fragment of...
| Supramolecule | Name: Purified Lassa virus VLPs at pH 5 mixed with purified fragment of LAMP1 type: sample / ID: 1000 / Details: Unfixed virus-like particles / Number unique components: 2 |
|---|
-Supramolecule #1: Lassa virus
| Supramolecule | Name: Lassa virus / type: virus / ID: 1 / Name.synonym: Lassa mammarenavirus / NCBI-ID: 11620 / Sci species name: Lassa virus / Sci species strain: Josiah / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No / Syn species name: Lassa mammarenavirus |
|---|---|
| Host (natural) | Organism:  Mastomys (multimammate rats) / synonym: VERTEBRATES Mastomys (multimammate rats) / synonym: VERTEBRATES |
| Host system | Organism:  |
-Macromolecule #1: lysosome-associated membrane glycoprotein 1
| Macromolecule | Name: lysosome-associated membrane glycoprotein 1 / type: protein_or_peptide / ID: 1 Name.synonym: lysosomal-associated membrane protein 1, LAMP1 Number of copies: 3 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Location in cell: Lysosome Homo sapiens (human) / synonym: Human / Location in cell: Lysosome |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293T Homo sapiens (human) / Recombinant cell: HEK293T |
| Sequence | UniProtKB: Lysosome-associated membrane glycoprotein 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 5.5 Details: 50 mM buffer of succinic acid, dihydrogen phosphate and glycine (2:7:7) |
|---|---|
| Grid | Details: Grids (Cflat CF-2/1-2C-T) were glow-discharged for 15 s. 6-nm gold particles were added. |
| Vitrification | Cryogen name: ETHANE-PROPANE MIXTURE / Chamber humidity: 80 % / Chamber temperature: 120 K / Instrument: GATAN CRYOPLUNGE 3 / Method: Blot for 3 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 80 K / Max: 120 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 160,000 times magnification. |
| Specialist optics | Energy filter - Name: GIF QUANTUM LS / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Details | Super-resolution counting mode |
| Date | Sep 25, 2015 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Digitization - Sampling interval: 5 µm / Number real images: 30 / Average electron dose: 60 e/Å2 Details: Each image is a tilt series of 19 movies, acquired at 5 degree intervals. Each movie consists of 8 frames. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 37037 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 4.6 µm / Nominal defocus min: 1.4 µm / Nominal magnification: 160000 |
| Sample stage | Specimen holder: Liquid nitrogen cooled / Specimen holder model: OTHER / Tilt series - Axis1 - Min angle: -45 ° / Tilt series - Axis1 - Max angle: 45 ° |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















