+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Native Photosystem I of Chlamydomonas reinhardtii | |||||||||
 Map data Map data | Native Photosystem I of Chlamydomonas reinhardtii | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | membrane supercomplex / light harvesting / electron transport / green algae / PHOTOSYNTHESIS | |||||||||
| Function / homology |  Function and homology information Function and homology informationchloroplast thylakoid lumen / photosynthesis, light harvesting / photosynthesis, light harvesting in photosystem I / photosystem I reaction center / photosystem I / photosynthetic electron transport in photosystem I / photosystem I / photosystem II / chloroplast thylakoid membrane / chlorophyll binding ...chloroplast thylakoid lumen / photosynthesis, light harvesting / photosynthesis, light harvesting in photosystem I / photosystem I reaction center / photosystem I / photosynthetic electron transport in photosystem I / photosystem I / photosystem II / chloroplast thylakoid membrane / chlorophyll binding / response to light stimulus / photosynthesis / 4 iron, 4 sulfur cluster binding / electron transfer activity / oxidoreductase activity / magnesium ion binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
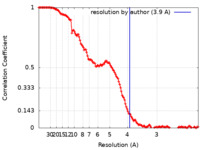
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Kurisu G / Gerle C | |||||||||
| Funding support |  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: Biochim Biophys Acta Bioenerg / Year: 2023 Journal: Biochim Biophys Acta Bioenerg / Year: 2023Title: Three structures of PSI-LHCI from Chlamydomonas reinhardtii suggest a resting state re-activated by ferredoxin. Authors: Christoph Gerle / Yuko Misumi / Akihiro Kawamoto / Hideaki Tanaka / Hisako Kubota-Kawai / Ryutaro Tokutsu / Eunchul Kim / Dror Chorev / Kazuhiro Abe / Carol V Robinson / Kaoru Mitsuoka / Jun ...Authors: Christoph Gerle / Yuko Misumi / Akihiro Kawamoto / Hideaki Tanaka / Hisako Kubota-Kawai / Ryutaro Tokutsu / Eunchul Kim / Dror Chorev / Kazuhiro Abe / Carol V Robinson / Kaoru Mitsuoka / Jun Minagawa / Genji Kurisu /   Abstract: Photosystem I (PSI) from the green alga Chlamydomonas reinhardtii, with various numbers of membrane bound antenna complexes (LHCI), has been described in great detail. In contrast, structural ...Photosystem I (PSI) from the green alga Chlamydomonas reinhardtii, with various numbers of membrane bound antenna complexes (LHCI), has been described in great detail. In contrast, structural characterization of soluble binding partners is less advanced. Here, we used X-ray crystallography and single particle cryo-EM to investigate three structures of the PSI-LHCI supercomplex from Chlamydomonas reinhardtii. An X-ray structure demonstrates the absence of six chlorophylls from the luminal side of the LHCI belts, suggesting these pigments were either physically absent or less stably associated with the complex, potentially influencing excitation transfer significantly. CryoEM revealed extra densities on luminal and stromal sides of the supercomplex, situated in the vicinity of the electron transfer sites. These densities disappeared after the binding of oxidized ferredoxin to PSI-LHCI. Based on these structures, we propose the existence of a PSI-LHCI resting state with a reduced active chlorophyll content, electron donors docked in waiting positions and regulatory binding partners positioned at the electron acceptor site. The resting state PSI-LHCI supercomplex would be recruited to its active form by the availability of oxidized ferredoxin. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32892.map.gz emd_32892.map.gz | 217.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32892-v30.xml emd-32892-v30.xml emd-32892.xml emd-32892.xml | 32.7 KB 32.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32892_fsc.xml emd_32892_fsc.xml | 13.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_32892.png emd_32892.png | 55 KB | ||
| Filedesc metadata |  emd-32892.cif.gz emd-32892.cif.gz | 9.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32892 http://ftp.pdbj.org/pub/emdb/structures/EMD-32892 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32892 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32892 | HTTPS FTP |
-Validation report
| Summary document |  emd_32892_validation.pdf.gz emd_32892_validation.pdf.gz | 540.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32892_full_validation.pdf.gz emd_32892_full_validation.pdf.gz | 540.3 KB | Display | |
| Data in XML |  emd_32892_validation.xml.gz emd_32892_validation.xml.gz | 13.8 KB | Display | |
| Data in CIF |  emd_32892_validation.cif.gz emd_32892_validation.cif.gz | 18.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32892 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32892 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32892 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32892 | HTTPS FTP |
-Related structure data
| Related structure data |  7wyiMC  7wznC  8h2uC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32892.map.gz / Format: CCP4 / Size: 229.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32892.map.gz / Format: CCP4 / Size: 229.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Native Photosystem I of Chlamydomonas reinhardtii | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
+Entire : Native Photosystem I with bound Light Harvesting Complex I from C...
+Supramolecule #1: Native Photosystem I with bound Light Harvesting Complex I from C...
+Macromolecule #1: Photosystem I P700 chlorophyll a apoprotein A1
+Macromolecule #2: Photosystem I P700 chlorophyll a apoprotein A2
+Macromolecule #3: Photosystem I iron-sulfur center
+Macromolecule #4: Photosystem I reaction center subunit II, chloroplastic
+Macromolecule #5: Photosystem I reaction center subunit IV, chloroplastic
+Macromolecule #6: Photosystem I reaction center subunit III, chloroplastic
+Macromolecule #7: Photosystem I reaction center subunit IX
+Macromolecule #8: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #9: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #10: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #11: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #12: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #13: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #14: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #15: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #16: CHLOROPHYLL A ISOMER
+Macromolecule #17: CHLOROPHYLL A
+Macromolecule #18: PHYLLOQUINONE
+Macromolecule #19: IRON/SULFUR CLUSTER
+Macromolecule #20: CHLOROPHYLL B
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 7.0 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: MOLYBDENUM / Mesh: 200 Details: Both sides of the grid were glow discharged at 5 mA for 90 seconds. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: 2.6 microliter of sample were applied to cryo-grids and blotted for 4 seconds using Whatman #1 before plunge freezing.. | |||||||||
| Details | The sample was monodisperse with a low level of contaminants and excess free detergent removed using the GraDeR approach. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | Preliminary grid screening was performed manually using a JEOL JEM-3000SFF microscope. |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Frames/image: 1-16 / Number grids imaged: 1 / Number real images: 4080 / Average exposure time: 2.0 sec. / Average electron dose: 45.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.25 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)