Entry Database : EMDB / ID : EMD-32735Title Structure of Human IGF1/IGFBP3/ALS Ternary Complex Complex : Ternary complex of IGF1/IGFBP3/ALSProtein or peptide : Insulin-like growth factor-binding protein complex acid labile subunitProtein or peptide : Insulin-like growth factor-binding protein 3Protein or peptide : Isoform 3 of Insulin-like growth factor ILigand : 2-acetamido-2-deoxy-beta-D-glucopyranose / / / / Function / homology Function Domain/homology Component
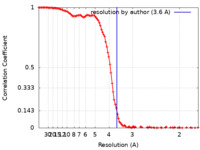
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / Resolution : 3.6 Å Kim H / Fu Y / Kim HM Funding support 1 items Organization Grant number Country Other government Institure of Basic Science, IBS-R030-C1
Journal : Nat Commun / Year : 2022Title : Structural basis for assembly and disassembly of the IGF/IGFBP/ALS ternary complexAuthors : Kim H / Fu Y / Hong HJ / Lee SG / Lee DS / Kim HM History Deposition Jan 27, 2022 - Header (metadata) release Aug 10, 2022 - Map release Aug 10, 2022 - Update Oct 23, 2024 - Current status Oct 23, 2024 Processing site : PDBj / Status : Released
Show all Show less
 Open data
Open data Basic information
Basic information
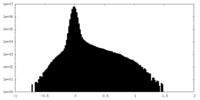
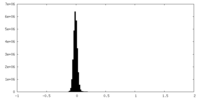
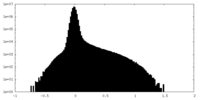
 Map data
Map data Sample
Sample Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) Authors
Authors Citation
Citation Journal: Nat Commun / Year: 2022
Journal: Nat Commun / Year: 2022 Structure visualization
Structure visualization Downloads & links
Downloads & links emd_32735.map.gz
emd_32735.map.gz EMDB map data format
EMDB map data format emd-32735-v30.xml
emd-32735-v30.xml emd-32735.xml
emd-32735.xml EMDB header
EMDB header emd_32735_fsc.xml
emd_32735_fsc.xml FSC data file
FSC data file emd_32735.png
emd_32735.png emd_32735_msk_1.map
emd_32735_msk_1.map Mask map
Mask map emd-32735.cif.gz
emd-32735.cif.gz emd_32735_half_map_1.map.gz
emd_32735_half_map_1.map.gz emd_32735_half_map_2.map.gz
emd_32735_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-32735
http://ftp.pdbj.org/pub/emdb/structures/EMD-32735 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32735
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32735 emd_32735_validation.pdf.gz
emd_32735_validation.pdf.gz EMDB validaton report
EMDB validaton report emd_32735_full_validation.pdf.gz
emd_32735_full_validation.pdf.gz emd_32735_validation.xml.gz
emd_32735_validation.xml.gz emd_32735_validation.cif.gz
emd_32735_validation.cif.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32735
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32735 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32735
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32735
 F&H Search
F&H Search Links
Links EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource Map
Map Download / File: emd_32735.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Download / File: emd_32735.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) emd_32735_msk_1.map
emd_32735_msk_1.map Sample components
Sample components Homo sapiens (human)
Homo sapiens (human) Homo sapiens (human)
Homo sapiens (human) Homo sapiens (human)
Homo sapiens (human) Homo sapiens (human)
Homo sapiens (human) Homo sapiens (human)
Homo sapiens (human) Homo sapiens (human)
Homo sapiens (human) Homo sapiens (human)
Homo sapiens (human)
 Processing
Processing Sample preparation
Sample preparation Electron microscopy
Electron microscopy FIELD EMISSION GUN
FIELD EMISSION GUN
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)