[English] 日本語
 Yorodumi
Yorodumi- EMDB-32631: Cryo-EM structure of ATP synthase dimer from Tetrahymena thermophila -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of ATP synthase dimer from Tetrahymena thermophila | |||||||||
 Map data Map data | Overall refinement of Tetrahymena thermophila ATP synthase dimer with C1 symmetry | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mitochondrial respiration / electron transport chain / ATP synthase / cristae / membrane complex / ELECTRON TRANSPORT | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.92 Å | |||||||||
 Authors Authors | Zhou L / Maldonado M / Padavannil A / Guo F / Letts JA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Structures of 's respiratory chain reveal the diversity of eukaryotic core metabolism. Authors: Long Zhou / María Maldonado / Abhilash Padavannil / Fei Guo / James A Letts /   Abstract: Respiration is a core biological energy-converting process whose last steps are carried out by a chain of multisubunit complexes in the inner mitochondrial membrane. To probe the functional and ...Respiration is a core biological energy-converting process whose last steps are carried out by a chain of multisubunit complexes in the inner mitochondrial membrane. To probe the functional and structural diversity of eukaryotic respiration, we examined the respiratory chain of the ciliate (Tt). Using cryo-electron microscopy on a mixed sample, we solved structures of a supercomplex between Tt complex I (Tt-CI) and Tt-CIII (Tt-SC I+III) and a structure of Tt-CIV. Tt-SC I+III (~2.3 megadaltons) is a curved assembly with structural and functional symmetry breaking. Tt-CIV is a ~2.7-megadalton dimer with more than 50 subunits per protomer, including mitochondrial carriers and a TIM8-TIM13-like domain. Our structural and functional study of the respiratory chain reveals divergence in key components of eukaryotic respiration, thereby expanding our understanding of core metabolism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32631.map.gz emd_32631.map.gz | 777.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32631-v30.xml emd-32631-v30.xml emd-32631.xml emd-32631.xml | 13.6 KB 13.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32631_fsc.xml emd_32631_fsc.xml | 20.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_32631.png emd_32631.png | 68 KB | ||
| Filedesc metadata |  emd-32631.cif.gz emd-32631.cif.gz | 4.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32631 http://ftp.pdbj.org/pub/emdb/structures/EMD-32631 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32631 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32631 | HTTPS FTP |
-Validation report
| Summary document |  emd_32631_validation.pdf.gz emd_32631_validation.pdf.gz | 589 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32631_full_validation.pdf.gz emd_32631_full_validation.pdf.gz | 588.6 KB | Display | |
| Data in XML |  emd_32631_validation.xml.gz emd_32631_validation.xml.gz | 17.1 KB | Display | |
| Data in CIF |  emd_32631_validation.cif.gz emd_32631_validation.cif.gz | 23.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32631 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32631 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32631 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32631 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32631.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32631.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Overall refinement of Tetrahymena thermophila ATP synthase dimer with C1 symmetry | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||||||||||||||||||
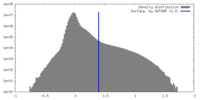
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Tetrahymena thermophila ATP synthase dimer
| Entire | Name: Tetrahymena thermophila ATP synthase dimer |
|---|---|
| Components |
|
-Supramolecule #1: Tetrahymena thermophila ATP synthase dimer
| Supramolecule | Name: Tetrahymena thermophila ATP synthase dimer / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.0 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: Solutions were made fresh from concentrated stocks, filtered and de-gassed before equilibration onto Superose6 column | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Details: 30 mA | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK III Details: incubation time before blotting: 60s blot time: 9s blot force: 25 offset: -2 incubation after blotting: 0s. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 7895 / Average exposure time: 5.9 sec. / Average electron dose: 66.69 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 58616 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)