+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of Leishmanial GDP-mannose pyrophosphorylase | |||||||||
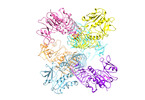
 Map data Map data | Apo LdGDP-MP | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information mannose-1-phosphate guanylyltransferase / mannose-1-phosphate guanylyltransferase (GTP) activity / GDP-mannose biosynthetic process / protein glycosylation / GTP binding / mannose-1-phosphate guanylyltransferase / mannose-1-phosphate guanylyltransferase (GTP) activity / GDP-mannose biosynthetic process / protein glycosylation / GTP binding /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Leishmania donovani (eukaryote) Leishmania donovani (eukaryote) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.4 Å cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Xu W / Li H / Huang C | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2022 Journal: Cell Discov / Year: 2022Title: Structural insights into selective inhibition of leishmanial GDP-mannose pyrophosphorylase. Authors: Hang Li / Tuo Ji / Qi Sun / Yao Chen / Weiya Xu / Chengdong Huang /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32509.map.gz emd_32509.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32509-v30.xml emd-32509-v30.xml emd-32509.xml emd-32509.xml | 9.4 KB 9.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32509.png emd_32509.png | 80.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32509 http://ftp.pdbj.org/pub/emdb/structures/EMD-32509 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32509 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32509 | HTTPS FTP |
-Related structure data
| Related structure data |  7whrMC  7whsC  7whtC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32509.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32509.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Apo LdGDP-MP | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.01 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Ld GDP-MP
| Entire | Name: Ld GDP-MP |
|---|---|
| Components |
|
-Supramolecule #1: Ld GDP-MP
| Supramolecule | Name: Ld GDP-MP / type: cell / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Leishmania donovani (eukaryote) Leishmania donovani (eukaryote) |
-Macromolecule #1: Nucleotidyl transferase family protein
| Macromolecule | Name: Nucleotidyl transferase family protein / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Leishmania donovani (eukaryote) Leishmania donovani (eukaryote) |
| Molecular weight | Theoretical: 41.780074 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MSASDGQGMR AVILVGGFGT RLRPLTLTTP KPLVPFCNKP MIIHQIEALK AVGVTEVILA VAYRPEAMKE QMDEWSRKLG VSFVFSVEE EPLGTAGPLA LARDILMQDD KPFFVLNSDV TCTFPMQELL DFHKAHGGEG TIMVSQVTQW EKYGVVVYSP Q NYQIERFV ...String: MSASDGQGMR AVILVGGFGT RLRPLTLTTP KPLVPFCNKP MIIHQIEALK AVGVTEVILA VAYRPEAMKE QMDEWSRKLG VSFVFSVEE EPLGTAGPLA LARDILMQDD KPFFVLNSDV TCTFPMQELL DFHKAHGGEG TIMVSQVTQW EKYGVVVYSP Q NYQIERFV EKPSRFLGDR INAGIYIFNK SILDRIPPRR ASIEKEIFPA MAAEGQLYAF NLEGFWMDVG QPKDYILGMT KF IPSLVHG NRETEQLHTE AVEHQRGGRF TVIGASLIDP SAKIGDGAVI GPYASIGANC VIGESCRIDN AAILENSKVG KGT MVSRSI VGWNNRIGSW CHIKDISVLG DDVEVKDGVI LIGTKVLPNK DVGEHRFEPG IIM |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 1.6 µm Bright-field microscopy / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 1.6 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 51.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
|---|---|
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 56825 |
 Movie
Movie Controller
Controller