[English] 日本語
 Yorodumi
Yorodumi- EMDB-32294: Plant receptor like protein RXEG1 in complex with xyloglucanase XEG1 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Plant receptor like protein RXEG1 in complex with xyloglucanase XEG1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to other organism / cellulase activity / polysaccharide catabolic process / defense response / membrane Similarity search - Function | |||||||||
| Biological species |  Phytophthora sojae (eukaryote) / Phytophthora sojae (eukaryote) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.11 Å | |||||||||
 Authors Authors | Sun Y / Wang Y / Zhang XX / Chen ZD / Xia YQ / Sun YJ / Zhang MM / Xiao Y / Han ZF / Wang YC / Chai JJ | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Plant receptor-like protein activation by a microbial glycoside hydrolase. Authors: Yue Sun / Yan Wang / Xiaoxiao Zhang / Zhaodan Chen / Yeqiang Xia / Lei Wang / Yujing Sun / Mingmei Zhang / Yu Xiao / Zhifu Han / Yuanchao Wang / Jijie Chai /   Abstract: Plants rely on cell-surface-localized pattern recognition receptors to detect pathogen- or host-derived danger signals and trigger an immune response. Receptor-like proteins (RLPs) with a leucine- ...Plants rely on cell-surface-localized pattern recognition receptors to detect pathogen- or host-derived danger signals and trigger an immune response. Receptor-like proteins (RLPs) with a leucine-rich repeat (LRR) ectodomain constitute a subgroup of pattern recognition receptors and play a critical role in plant immunity. Mechanisms underlying ligand recognition and activation of LRR-RLPs remain elusive. Here we report a crystal structure of the LRR-RLP RXEG1 from Nicotiana benthamiana that recognizes XEG1 xyloglucanase from the pathogen Phytophthora sojae. The structure reveals that specific XEG1 recognition is predominantly mediated by an amino-terminal and a carboxy-terminal loop-out region (RXEG1(ID)) of RXEG1. The two loops bind to the active-site groove of XEG1, inhibiting its enzymatic activity and suppressing Phytophthora infection of N. benthamiana. Binding of XEG1 promotes association of RXEG1(LRR) with the LRR-type co-receptor BAK1 through RXEG1(ID) and the last four conserved LRRs to trigger RXEG1-mediated immune responses. Comparison of the structures of apo-RXEG1(LRR), XEG1-RXEG1(LRR) and XEG1-BAK1-RXEG1(LRR) shows that binding of XEG1 induces conformational changes in the N-terminal region of RXEG1(ID) and enhances structural flexibility of the BAK1-associating regions of RXEG1(LRR). These changes allow fold switching of RXEG1(ID) for recruitment of BAK1(LRR). Our data reveal a conserved mechanism of ligand-induced heterodimerization of an LRR-RLP with BAK1 and suggest a dual function for the LRR-RLP in plant immunity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32294.map.gz emd_32294.map.gz | 47.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32294-v30.xml emd-32294-v30.xml emd-32294.xml emd-32294.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32294.png emd_32294.png | 20.6 KB | ||
| Others |  emd_32294_half_map_1.map.gz emd_32294_half_map_1.map.gz emd_32294_half_map_2.map.gz emd_32294_half_map_2.map.gz | 40.9 MB 40.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32294 http://ftp.pdbj.org/pub/emdb/structures/EMD-32294 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32294 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32294 | HTTPS FTP |
-Related structure data
| Related structure data |  7w3vMC  7drbC  7drcC  7w3tC  7w3xC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32294.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32294.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.061 Å | ||||||||||||||||||||||||||||||||||||
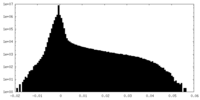
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_32294_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
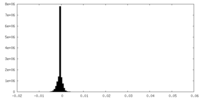
| Density Histograms |
-Half map: #2
| File | emd_32294_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
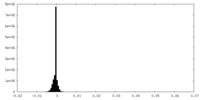
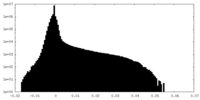
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of XEG1 bounded RXEG1
| Entire | Name: Ternary complex of XEG1 bounded RXEG1 |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of XEG1 bounded RXEG1
| Supramolecule | Name: Ternary complex of XEG1 bounded RXEG1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|
-Supramolecule #2: XEG1
| Supramolecule | Name: XEG1 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Phytophthora sojae (eukaryote) Phytophthora sojae (eukaryote) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
-Supramolecule #3: RXEG1
| Supramolecule | Name: RXEG1 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
-Macromolecule #1: Cell 12A endoglucanase
| Macromolecule | Name: Cell 12A endoglucanase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Phytophthora sojae (eukaryote) Phytophthora sojae (eukaryote) |
| Molecular weight | Theoretical: 25.519551 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MKGFFAGVVA AATLAVASAG DYCGQWDWAK STNYIVYNNL WNKNAAASGS QCTGVDKISG STIAWHTSYT WTGGAATEVK SYSNAALVF SKKQIKNIKS IPTKMKYSYS HSSGTFVADV SYDLFTSSTA SGSNEYEIMI WLAAYGGAGP ISSTGKAIAT V TIGSNSFK ...String: MKGFFAGVVA AATLAVASAG DYCGQWDWAK STNYIVYNNL WNKNAAASGS QCTGVDKISG STIAWHTSYT WTGGAATEVK SYSNAALVF SKKQIKNIKS IPTKMKYSYS HSSGTFVADV SYDLFTSSTA SGSNEYEIMI WLAAYGGAGP ISSTGKAIAT V TIGSNSFK LYKGPNGSTT VFSFVATKTI TNFSADLQKF LSYLTKNQGL PSSQYLITLE AGTEPFVGTN AKMTVSSFSA AV N |
-Macromolecule #2: Membrane-localized LRR receptor-like protein
| Macromolecule | Name: Membrane-localized LRR receptor-like protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 104.238852 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGKREYPSSA HFLVTLSLLL LQAAFGLTLC IEKERDALLE FKRGLSDNFG QLSTWGDEED KKECCKWKGI ECNKTTGHVI VLDLHNAFT CSASACFAPR LTGKLSPSLL ELEYLNFLDL SVNEFERSEI PRFICSFKRL EYLNLSSSFF SGLIPTQFKN L TSLRILDL ...String: MGKREYPSSA HFLVTLSLLL LQAAFGLTLC IEKERDALLE FKRGLSDNFG QLSTWGDEED KKECCKWKGI ECNKTTGHVI VLDLHNAFT CSASACFAPR LTGKLSPSLL ELEYLNFLDL SVNEFERSEI PRFICSFKRL EYLNLSSSFF SGLIPTQFKN L TSLRILDL GYNNLIVKDL TWLSHLSSLE LLSLGGSDFQ VKNWFQEITK LPLLKELDLS LCGLSKLVPS PAEIANSSLI SL SVLHLCC NEFSSSAKYS WLFNFSTSLT SIDLSNNQLD GQIDDRFGNL MYLEHLNLAN ELNLKGGIPS SFGNLTRLRY LDM SNTRTY QWLPELFVRL SGSRKTLEVL GLNDNSMFGS LVDVTRFSAL KRLYLQKNVL NGFFMERFGQ VSSLEYLDLS DNQM RGPLP DLALFPSLRE LHLGSNHFNG RIPQGIGKLS QLKILDVSSN RLEGLPESMG QLSNLESFDA SYNVLKGTIT ESHLS NLSS LVDLDLSFNS LALKTSIDWL PPFQLQVINL PSCNLGPSFP KWLQSQNNYT VLDISLANIS DALPSWFSGL PPDIKI LNL SNNQISGRVS DLIENAYDYM VIDLSSNNFS GPLPLVPTNV QIFYLHKNQF FGSISSICKS TTGATSLDLS HNQFSGE LP DCWMNATNLA VLNLAYNNFS GKLPQSLGSL TNLEALYMRQ NSFSGMLPSL SQCQSLQILD LGGNKLTGRI PAWIGTDL L NLRILSLRFN KFYGSISPII CQLQFLQILD LSANGLAGKI PQCFNNFTLL HQENGLGEPM EFLVQGFYGK YPRHYSYLG NLLVQWKNQE AEYKNPLTYL KTIDLSSNKL VGGIPKEMAE MRGLKSLNLS RNDLNGSIIK GIGQMKMLES LDLSRNQLSG MIPKDLANL TFIGVLDLSN NHLSGRIPSS TQLQTFERSS YSGNAQLCGP PLQEC |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 8 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.11 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 300 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)