[English] 日本語
 Yorodumi
Yorodumi- EMDB-32293: Cryo-EM structure of plant receptor like kinase NbBAK1 in RXEG1-B... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of plant receptor like kinase NbBAK1 in RXEG1-BAK1-XEG1 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | LRR / PTI / Glycoside hydrolase / Inhibitor / PLANT PROTEIN | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.59 Å | |||||||||
 Authors Authors | Sun Y / Wang Y / Zhang XX / Chen ZD / Xia YQ / Sun YJ / Zhang MM / Xiao Y / Han ZF / Wang YC / Chai JJ | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Plant receptor-like protein activation by a microbial glycoside hydrolase. Authors: Yue Sun / Yan Wang / Xiaoxiao Zhang / Zhaodan Chen / Yeqiang Xia / Lei Wang / Yujing Sun / Mingmei Zhang / Yu Xiao / Zhifu Han / Yuanchao Wang / Jijie Chai /   Abstract: Plants rely on cell-surface-localized pattern recognition receptors to detect pathogen- or host-derived danger signals and trigger an immune response. Receptor-like proteins (RLPs) with a leucine- ...Plants rely on cell-surface-localized pattern recognition receptors to detect pathogen- or host-derived danger signals and trigger an immune response. Receptor-like proteins (RLPs) with a leucine-rich repeat (LRR) ectodomain constitute a subgroup of pattern recognition receptors and play a critical role in plant immunity. Mechanisms underlying ligand recognition and activation of LRR-RLPs remain elusive. Here we report a crystal structure of the LRR-RLP RXEG1 from Nicotiana benthamiana that recognizes XEG1 xyloglucanase from the pathogen Phytophthora sojae. The structure reveals that specific XEG1 recognition is predominantly mediated by an amino-terminal and a carboxy-terminal loop-out region (RXEG1(ID)) of RXEG1. The two loops bind to the active-site groove of XEG1, inhibiting its enzymatic activity and suppressing Phytophthora infection of N. benthamiana. Binding of XEG1 promotes association of RXEG1(LRR) with the LRR-type co-receptor BAK1 through RXEG1(ID) and the last four conserved LRRs to trigger RXEG1-mediated immune responses. Comparison of the structures of apo-RXEG1(LRR), XEG1-RXEG1(LRR) and XEG1-BAK1-RXEG1(LRR) shows that binding of XEG1 induces conformational changes in the N-terminal region of RXEG1(ID) and enhances structural flexibility of the BAK1-associating regions of RXEG1(LRR). These changes allow fold switching of RXEG1(ID) for recruitment of BAK1(LRR). Our data reveal a conserved mechanism of ligand-induced heterodimerization of an LRR-RLP with BAK1 and suggest a dual function for the LRR-RLP in plant immunity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32293.map.gz emd_32293.map.gz | 46.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32293-v30.xml emd-32293-v30.xml emd-32293.xml emd-32293.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32293.png emd_32293.png | 5.8 KB | ||
| Filedesc metadata |  emd-32293.cif.gz emd-32293.cif.gz | 6.1 KB | ||
| Others |  emd_32293_half_map_1.map.gz emd_32293_half_map_1.map.gz emd_32293_half_map_2.map.gz emd_32293_half_map_2.map.gz | 40.8 MB 40.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32293 http://ftp.pdbj.org/pub/emdb/structures/EMD-32293 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32293 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32293 | HTTPS FTP |
-Related structure data
| Related structure data |  7w3tMC  7drbC  7drcC  7w3vC  7w3xC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32293.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32293.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.061 Å | ||||||||||||||||||||||||||||||||||||
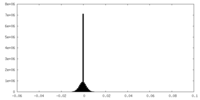
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_32293_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
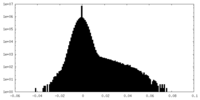
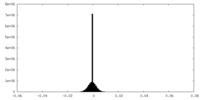
| Density Histograms |
-Half map: #2
| File | emd_32293_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
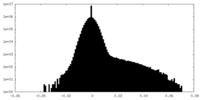
| Density Histograms |
- Sample components
Sample components
-Entire : BAK1
| Entire | Name: BAK1 |
|---|---|
| Components |
|
-Supramolecule #1: BAK1
| Supramolecule | Name: BAK1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Brassinosteroid insensitive 1-associated receptor kinase 1
| Macromolecule | Name: Brassinosteroid insensitive 1-associated receptor kinase 1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.514012 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MDQWILGILG FVSTFLCLIG LLLVPVSANI EGDALNALKT NLADPNNVLQ SWDPTLVNPC TWFHVTCNSE NSVTRVDLGN ANLSGQLVP QLGQLPNLQY LELYSNNISG RIPFELGNLT NLVSLDLYLN RLNGPIPDTL GKLQKLRFLR LNNNSLNGRI P MLLTTVIS ...String: MDQWILGILG FVSTFLCLIG LLLVPVSANI EGDALNALKT NLADPNNVLQ SWDPTLVNPC TWFHVTCNSE NSVTRVDLGN ANLSGQLVP QLGQLPNLQY LELYSNNISG RIPFELGNLT NLVSLDLYLN RLNGPIPDTL GKLQKLRFLR LNNNSLNGRI P MLLTTVIS LQVLDLSNNN LTGPVPVNGS FSLFTPISFA NNPLDIPPAA PPPPISPMPP SSSGVGNSAT |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)