[English] 日本語
 Yorodumi
Yorodumi- EMDB-31843: Nucleotide-free V1EG domain of V/A-ATPase from Thermus thermophil... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Nucleotide-free V1EG domain of V/A-ATPase from Thermus thermophilus, state1-2 | |||||||||||||||||||||
 Map data Map data | Nucleotide-free V1EG domain of V/A-ATPase from Thermus thermophilus, state1-2, postprocessed map. | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | rotary ATPase / V-type ATPase / ATP synthase / Thermus thermophilus / chemo-mechanical coupling / MOTOR PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationproton-transporting two-sector ATPase complex, catalytic domain / proton-transporting ATP synthase complex / proton motive force-driven plasma membrane ATP synthesis / H+-transporting two-sector ATPase / proton-transporting ATPase activity, rotational mechanism / proton-transporting ATP synthase activity, rotational mechanism / ATP binding / metal ion binding Similarity search - Function | |||||||||||||||||||||
| Biological species |   Thermus thermophilus HB8 (bacteria) Thermus thermophilus HB8 (bacteria) | |||||||||||||||||||||
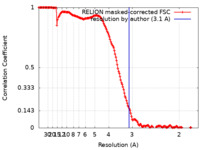
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||||||||||||||
 Authors Authors | Kishikawa J / Nakanishi A / Nakano A / Saeki S / Furuta A / Kato T / Mitsuoka K / Yokoyama K | |||||||||||||||||||||
| Funding support |  Japan, 6 items Japan, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases. Authors: J Kishikawa / A Nakanishi / A Nakano / S Saeki / A Furuta / T Kato / K Mistuoka / K Yokoyama /  Abstract: V/A-ATPase is a motor protein that shares a common rotary catalytic mechanism with FF ATP synthase. When powered by ATP hydrolysis, the V domain rotates the central rotor against the AB hexamer, ...V/A-ATPase is a motor protein that shares a common rotary catalytic mechanism with FF ATP synthase. When powered by ATP hydrolysis, the V domain rotates the central rotor against the AB hexamer, composed of three catalytic AB dimers adopting different conformations (AB, AB, and AB). Here, we report the atomic models of 18 catalytic intermediates of the V domain of V/A-ATPase under different reaction conditions, determined by single particle cryo-EM. The models reveal that the rotor does not rotate immediately after binding of ATP to the V. Instead, three events proceed simultaneously with the 120˚ rotation of the shaft: hydrolysis of ATP in AB, zipper movement in AB by the binding ATP, and unzipper movement in AB with release of both ADP and Pi. This indicates the unidirectional rotation of V/A-ATPase by a ratchet-like mechanism owing to ATP hydrolysis in AB, rather than the power stroke model proposed previously for F-ATPase. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31843.map.gz emd_31843.map.gz | 326 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31843-v30.xml emd-31843-v30.xml emd-31843.xml emd-31843.xml | 24.1 KB 24.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31843_fsc.xml emd_31843_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_31843.png emd_31843.png | 115.9 KB | ||
| Masks |  emd_31843_msk_1.map emd_31843_msk_1.map | 347.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-31843.cif.gz emd-31843.cif.gz | 6.8 KB | ||
| Others |  emd_31843_half_map_1.map.gz emd_31843_half_map_1.map.gz emd_31843_half_map_2.map.gz emd_31843_half_map_2.map.gz | 278.9 MB 277.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31843 http://ftp.pdbj.org/pub/emdb/structures/EMD-31843 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31843 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31843 | HTTPS FTP |
-Validation report
| Summary document |  emd_31843_validation.pdf.gz emd_31843_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31843_full_validation.pdf.gz emd_31843_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_31843_validation.xml.gz emd_31843_validation.xml.gz | 23.3 KB | Display | |
| Data in CIF |  emd_31843_validation.cif.gz emd_31843_validation.cif.gz | 30.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31843 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31843 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31843 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31843 | HTTPS FTP |
-Related structure data
| Related structure data |  7vajMC  7vaiC  7vakC  7valC  7vamC  7vanC  7vaoC  7vapC  7vaqC  7varC  7vasC  7vatC  7vauC  7vavC  7vawC  7vaxC  7vayC  7vb0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31843.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31843.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Nucleotide-free V1EG domain of V/A-ATPase from Thermus thermophilus, state1-2, postprocessed map. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||||||||||||||||||
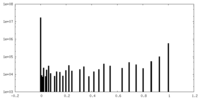
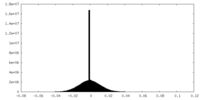
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_31843_msk_1.map emd_31843_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
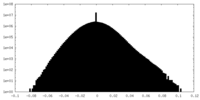
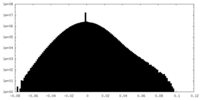
| Density Histograms |
-Half map: Nucleotide-free V1EG domain of V/A-ATPase from Thermus thermophilus,...
| File | emd_31843_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Nucleotide-free V1EG domain of V/A-ATPase from Thermus thermophilus, state1-2, halfmap2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Nucleotide-free V1EG domain of V/A-ATPase from Thermus thermophilus,...
| File | emd_31843_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Nucleotide-free V1EG domain of V/A-ATPase from Thermus thermophilus, state1-2, halfmap1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Nucleotide-free V1EG domain of V/A-ATPase from Thermus thermophil...
| Entire | Name: Nucleotide-free V1EG domain of V/A-ATPase from Thermus thermophilus, state1-2 |
|---|---|
| Components |
|
-Supramolecule #1: Nucleotide-free V1EG domain of V/A-ATPase from Thermus thermophil...
| Supramolecule | Name: Nucleotide-free V1EG domain of V/A-ATPase from Thermus thermophilus, state1-2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus HB8 (bacteria) Thermus thermophilus HB8 (bacteria) |
| Molecular weight | Theoretical: 500 KDa |
-Macromolecule #1: V-type ATP synthase alpha chain
| Macromolecule | Name: V-type ATP synthase alpha chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO / EC number: H+-transporting two-sector ATPase |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus HB8 (bacteria) / Strain: HB8 Thermus thermophilus HB8 (bacteria) / Strain: HB8 |
| Molecular weight | Theoretical: 63.669957 KDa |
| Sequence | String: MIQGVIQKIA GPAVIAKGML GARMYDICKV GEEGLVGEII RLDGDTAFVQ VYEDTSGLKV GEPVVSTGLP LAVELGPGML NGIYDGIQR PLERIREKTG IYITRGVVVH ALDREKKWAW TPMVKPGDEV RGGMVLGTVP EFGFTHKILV PPDVRGRVKE V KPAGEYTV ...String: MIQGVIQKIA GPAVIAKGML GARMYDICKV GEEGLVGEII RLDGDTAFVQ VYEDTSGLKV GEPVVSTGLP LAVELGPGML NGIYDGIQR PLERIREKTG IYITRGVVVH ALDREKKWAW TPMVKPGDEV RGGMVLGTVP EFGFTHKILV PPDVRGRVKE V KPAGEYTV EEPVVVLEDG TELKMYHTWP VRRARPVQRK LDPNTPFLTG MRILDVLFPV AMGGTAAIPG PFGAGKSVTQ QS LAKWSNA DVVVYVGCGE RGNEMTDVLV EFPELTDPKT GGPLMHRTVL IANTSNMPVA AREASIYVGV TIAEYFRDQG FSV ALMADS TSRWAEALRE ISSRLEEMPA EEGYPPYLAA RLAAFYERAG KVITLGGEEG AVTIVGAVSP PGGDMSEPVT QSTL RIVGA FWRLDASLAF RRHFPAINWN GSYSLFTSAL DPWYRENVAE DYPELRDAIS ELLQREAGLQ EIVQLVGPDA LQDAE RLVI EVGRIIREDF LQQNAYHEVD AYCSMKKAYG IMKMILAFYK EAEAAIKRGV SIDEILQLPV LERIGRARYV SEEEFP AYF EEAMKEIQGA FKALA UniProtKB: V-type ATP synthase alpha chain |
-Macromolecule #2: V-type ATP synthase beta chain
| Macromolecule | Name: V-type ATP synthase beta chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus HB8 (bacteria) / Strain: HB8 Thermus thermophilus HB8 (bacteria) / Strain: HB8 |
| Molecular weight | Theoretical: 53.2195 KDa |
| Sequence | String: MDLLKKEYTG ITYISGPLLF VENAKDLAYG AIVDIKDGTG RVRGGQVIEV SEEYAVIQVF EETTGLDLAT TSVSLVEDVA RLGVSKEML GRRFNGIGKP IDGLPPITPE KRLPITGLPL NPVARRKPEQ FIQTGISTID VMNTLVRGQK LPIFSGSGLP A NEIAAQIA ...String: MDLLKKEYTG ITYISGPLLF VENAKDLAYG AIVDIKDGTG RVRGGQVIEV SEEYAVIQVF EETTGLDLAT TSVSLVEDVA RLGVSKEML GRRFNGIGKP IDGLPPITPE KRLPITGLPL NPVARRKPEQ FIQTGISTID VMNTLVRGQK LPIFSGSGLP A NEIAAQIA RQATVRPDLS GEGEKEEPFA VVFAAMGITQ RELSYFIQEF ERTGALSRSV LFLNKADDPT IERILTPRMA LT VAEYLAF EHDYHVLVIL TDMTNYCEAL REIGAAREEI PGRRGYPGYM YTDLATIYER AGVVEGKKGS VTQIPILSMP DDD RTHPIP DLTGYITEGQ IQLSRELHRK GIYPPIDPLP SLSRLMNNGV GKGKTREDHK QVSDQLYSAY ANGVDIRKLV AIIG EDALT ENDRRYLQFA DAFERFFINQ GQQNRSIEES LQIAWALLSM LPQGELKRIS KDHIGKYYGQ KLEEIWGAPQ ALD UniProtKB: V-type ATP synthase beta chain |
-Macromolecule #3: V-type ATP synthase subunit D
| Macromolecule | Name: V-type ATP synthase subunit D / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus HB8 (bacteria) / Strain: HB8 Thermus thermophilus HB8 (bacteria) / Strain: HB8 |
| Molecular weight | Theoretical: 24.715566 KDa |
| Sequence | String: MSQVSPTRMN LLQRRGQLRL AQKGVDLLKK KRDALVAEFF GLVREAMEAR KALDQAAKEA YAALLLAQAF DGPEVVAGAA LGVPPLEGV EAEVENVWGS KVPRLKATFP DGALLSPVGT PAYTLEASRA FRRYAEALIR VANTETRLKK IGEEIKKTTR R VNALEQVV ...String: MSQVSPTRMN LLQRRGQLRL AQKGVDLLKK KRDALVAEFF GLVREAMEAR KALDQAAKEA YAALLLAQAF DGPEVVAGAA LGVPPLEGV EAEVENVWGS KVPRLKATFP DGALLSPVGT PAYTLEASRA FRRYAEALIR VANTETRLKK IGEEIKKTTR R VNALEQVV IPGIRAQIRF IQQVLEQRER EDTFRLKRIK GKIEAREAEE EGGRPNPQVE IGAGL UniProtKB: V-type ATP synthase subunit D |
-Macromolecule #4: V-type ATP synthase subunit F
| Macromolecule | Name: V-type ATP synthase subunit F / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus HB8 (bacteria) / Strain: HB8 Thermus thermophilus HB8 (bacteria) / Strain: HB8 |
| Molecular weight | Theoretical: 11.294904 KDa |
| Sequence | String: MAVIADPETA QGFRLAGLEG YGASSAEEAQ SLLETLVERG GYALVAVDEA LLPDPERAVE RLMRGRDLPV LLPIAGLKEA FQGHDVEGY MRELVRKTIG FDIKL UniProtKB: V-type ATP synthase subunit F |
-Macromolecule #5: V-type ATP synthase subunit G
| Macromolecule | Name: V-type ATP synthase subunit G / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus HB8 (bacteria) / Strain: HB8 Thermus thermophilus HB8 (bacteria) / Strain: HB8 |
| Molecular weight | Theoretical: 13.166218 KDa |
| Sequence | String: MTGGLVLNAI SRAGGAMGGL GLIKSLAEKE KQLLERLEAA KKEAEERVKR AEAEAKALLE EAEAKAKALE AQYRERERAE TEALLARYR ERAEAEAKAV REKAMARLDE AVALVLKEVL P UniProtKB: V-type ATP synthase, subunit (VAPC-THERM) |
-Macromolecule #6: V-type ATP synthase subunit E
| Macromolecule | Name: V-type ATP synthase subunit E / type: protein_or_peptide / ID: 6 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus HB8 (bacteria) / Strain: HB8 Thermus thermophilus HB8 (bacteria) / Strain: HB8 |
| Molecular weight | Theoretical: 20.645582 KDa |
| Sequence | String: MSKLEAILSQ EVEAEIQALL QEAEAKAEAV KREAEEKAKA LLQARERALE AQYRAALRRA ESAGELLVAT ARTQARGEVL EEVRRRVRE ALEALPQKPE WPEVVRKLAL EALEALPGAK ALVANPEDLP HLEALARERG VELQAEPALR LGVRAVGAEG K TQVENSLL ARLDRAWDAL SSKVAQALWG UniProtKB: V-type ATP synthase subunit E |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: MOLYBDENUM / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 5.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.045 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | The atomic model built in this study was used as an initial model. |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER |
| Output model |  PDB-7vaj: |
 Movie
Movie Controller
Controller






































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)