[English] 日本語
 Yorodumi
Yorodumi- EMDB-31776: Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31776 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 1 | |||||||||||||||
 Map data Map data | SARS-CoV-2 S-Delta variant, one RBD-up conformation 1, unsharpened map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationMaturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell ...Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / membrane fusion / receptor-mediated endocytosis of virus by host cell / Attachment and Entry / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / receptor ligand activity / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||||||||
 Authors Authors | Yang TJ / Yu PY / Chang YC / Hsu STD | |||||||||||||||
| Funding support |  Taiwan, 4 items Taiwan, 4 items
| |||||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 1 Authors: Yang TJ / Yu PY / Chang YC / Hsu STD | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31776.map.gz emd_31776.map.gz | 108.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31776-v30.xml emd-31776-v30.xml emd-31776.xml emd-31776.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31776.png emd_31776.png | 52.6 KB | ||
| Others |  emd_31776_additional_1.map.gz emd_31776_additional_1.map.gz | 203.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31776 http://ftp.pdbj.org/pub/emdb/structures/EMD-31776 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31776 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31776 | HTTPS FTP |
-Validation report
| Summary document |  emd_31776_validation.pdf.gz emd_31776_validation.pdf.gz | 433.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31776_full_validation.pdf.gz emd_31776_full_validation.pdf.gz | 432.8 KB | Display | |
| Data in XML |  emd_31776_validation.xml.gz emd_31776_validation.xml.gz | 7.2 KB | Display | |
| Data in CIF |  emd_31776_validation.cif.gz emd_31776_validation.cif.gz | 8.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31776 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31776 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31776 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31776 | HTTPS FTP |
-Related structure data
| Related structure data |  7v7oMC  7v76C  7v77C  7v78C  7v79C  7v7aC  7v7dC  7v7eC  7v7fC  7v7gC  7v7hC  7v7iC  7v7jC  7v7nC  7v7pC  7v7qC  7v7rC  7v7sC  7v7tC  7v7uC  7v7vC  7v7zC  7v80C  7v81C  7v82C  7v83C  7v84C  7v85C  7v86C  7v87C  7v88C  7v89C  7v8aC  7v8bC  7v8cC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31776.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31776.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-CoV-2 S-Delta variant, one RBD-up conformation 1, unsharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
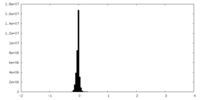
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: SARS-CoV-2 S-Delta variant, one RBD-up conformation 1, sharpened map
| File | emd_31776_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-CoV-2 S-Delta variant, one RBD-up conformation 1, sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
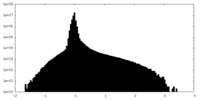
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 spike glycoprotein
| Entire | Name: SARS-CoV-2 spike glycoprotein |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 spike glycoprotein
| Supramolecule | Name: SARS-CoV-2 spike glycoprotein / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Delta variant (B.1.617.2), one RBD-up conformation 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 540 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 142.047203 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLRT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLDVYYHKNN KSWMESGVYS S ANNCTFEY ...String: MFVFLVLLPL VSSQCVNLRT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLDVYYHKNN KSWMESGVYS S ANNCTFEY VSQPFLMDLE GKQGNFKNLR EFVFKNIDGY FKIYSKHTPI NLVRDLPQGF SALEPLVDLP IGINITRFQT LL ALHRSYL TPGDSSSGWT AGAAAYYVGY LQPRTFLLKY NENGTITDAV DCALDPLSET KCTLKSFTVE KGIYQTSNFR VQP TESIVR FPNITNLCPF GEVFNATRFA SVYAWNRKRI SNCVADYSVL YNSASFSTFK CYGVSPTKLN DLCFTNVYAD SFVI RGDEV RQIAPGQTGK IADYNYKLPD DFTGCVIAWN SNNLDSKVGG NYNYRYRLFR KSNLKPFERD ISTEIYQAGS KPCNG VEGF NCYFPLQSYG FQPTNGVGYQ PYRVVVLSFE LLHAPATVCG PKKSTNLVKN KCVNFNFNGL TGTGVLTESN KKFLPF QQF GRDIADTTDA VRDPQTLEIL DITPCSFGGV SVITPGTNTS NQVAVLYQGV NCTEVPVAIH ADQLTPTWRV YSTGSNV FQ TRAGCLIGAE HVNNSYECDI PIGAGICASY QTQTNSRGSA SSVASQSIIA YTMSLGAENS VAYSNNSIAI PTNFTISV T TEILPVSMTK TSVDCTMYIC GDSTECSNLL LQYGSFCTQL NRALTGIAVE QDKNTQEVFA QVKQIYKTPP IKDFGGFNF SQILPDPSKP SKRSFIEDLL FNKVTLADAG FIKQYGDCLG DIAARDLICA QKFNGLTVLP PLLTDEMIAQ YTSALLAGTI TSGWTFGAG AALQIPFAMQ MAYRFNGIGV TQNVLYENQK LIANQFNSAI GKIQDSLSST ASALGKLQNV VNQNAQALNT L VKQLSSNF GAISSVLNDI LSRLDPPEAE VQIDRLITGR LQSLQTYVTQ QLIRAAEIRA SANLAATKMS ECVLGQSKRV DF CGKGYHL MSFPQSAPHG VVFLHVTYVP AQEKNFTTAP AICHDGKAHF PREGVFVSNG THWFVTQRNF YEPQIITTDN TFV SGNCDV VIGIVNNTVY DPLQPELDSF KEELDKYFKN HTSPDVDLGD ISGINASVVN IQKEIDRLNE VAKNLNESLI DLQE LGKYE QEFGSGGYIP EAPRDGQAYV RKDGEWVLLS TFLKGQDNSA DIQHSGRPLE SRGPFEQKLI SEEDLNMHTG HHHHH H |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 13 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: blot for 2.5 seconds before plunging; blot force: 0; waiting time: 30s. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)