[English] 日本語
 Yorodumi
Yorodumi- EMDB-3141: Electron cryo-microscopy of PrgJ/NAIP2/CARD-truncated NLRC4 infla... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3141 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron cryo-microscopy of PrgJ/NAIP2/CARD-truncated NLRC4 inflammasome with pseudo c11 symmetry | |||||||||
 Map data Map data | Reconstruction of PrgJ/NAIP2/CARD-truncated NLRC4 inflammasome with pseudo c11 symmetry | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | inflammasome / NLRC4 / NAIP | |||||||||
| Biological species |   Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) | |||||||||
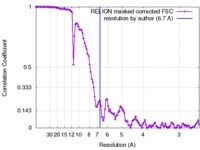
| Method | single particle reconstruction / cryo EM / Resolution: 6.7 Å | |||||||||
 Authors Authors | Hu ZH / Zhou Q / Zhang CL / Fan SL / Cheng W / Zhao Y / Shao F / Wang HW / Sui SF / Chai JJ | |||||||||
 Citation Citation |  Journal: Science / Year: 2015 Journal: Science / Year: 2015Title: Structural and biochemical basis for induced self-propagation of NLRC4. Authors: Zehan Hu / Qiang Zhou / Chenlu Zhang / Shilong Fan / Wei Cheng / Yue Zhao / Feng Shao / Hong-Wei Wang / Sen-Fang Sui / Jijie Chai /  Abstract: Responding to stimuli, nucleotide-binding domain and leucine-rich repeat-containing proteins (NLRs) oligomerize into multiprotein complexes, termed inflammasomes, mediating innate immunity. ...Responding to stimuli, nucleotide-binding domain and leucine-rich repeat-containing proteins (NLRs) oligomerize into multiprotein complexes, termed inflammasomes, mediating innate immunity. Recognition of bacterial pathogens by NLR apoptosis inhibitory proteins (NAIPs) induces NLR family CARD domain-containing protein 4 (NLRC4) activation and formation of NAIP-NLRC4 inflammasomes. The wheel-like structure of a PrgJ-NAIP2-NLRC4 complex determined by cryogenic electron microscopy at 6.6 angstrom reveals that NLRC4 activation involves substantial structural reorganization that creates one oligomerization surface (catalytic surface). Once activated, NLRC4 uses this surface to catalyze the activation of an inactive NLRC4, self-propagating its active conformation to form the wheel-like architecture. NAIP proteins possess a catalytic surface matching the other oligomerization surface (receptor surface) of NLRC4 but not those of their own, ensuring that one NAIP is sufficient to initiate NLRC4 oligomerization. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3141.map.gz emd_3141.map.gz | 114.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3141-v30.xml emd-3141-v30.xml emd-3141.xml emd-3141.xml | 11.5 KB 11.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3141_fsc.xml emd_3141_fsc.xml | 11.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_3141.png emd_3141.png | 393.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3141 http://ftp.pdbj.org/pub/emdb/structures/EMD-3141 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3141 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3141 | HTTPS FTP |
-Validation report
| Summary document |  emd_3141_validation.pdf.gz emd_3141_validation.pdf.gz | 289.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3141_full_validation.pdf.gz emd_3141_full_validation.pdf.gz | 288.9 KB | Display | |
| Data in XML |  emd_3141_validation.xml.gz emd_3141_validation.xml.gz | 12.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3141 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3141 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3141 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3141 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3141.map.gz / Format: CCP4 / Size: 122.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3141.map.gz / Format: CCP4 / Size: 122.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of PrgJ/NAIP2/CARD-truncated NLRC4 inflammasome with pseudo c11 symmetry | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.32 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : PrgJ/NAIP2/CARD-truncated NLRC4 inflammasome
| Entire | Name: PrgJ/NAIP2/CARD-truncated NLRC4 inflammasome |
|---|---|
| Components |
|
-Supramolecule #1000: PrgJ/NAIP2/CARD-truncated NLRC4 inflammasome
| Supramolecule | Name: PrgJ/NAIP2/CARD-truncated NLRC4 inflammasome / type: sample / ID: 1000 Oligomeric state: 11 heterotrimers of PrgJ, NAIP2 and CARD-truncated NLRC4 Number unique components: 3 |
|---|---|
| Molecular weight | Experimental: 1.1 MDa / Theoretical: 1.1 MDa |
-Macromolecule #1: NLRC4
| Macromolecule | Name: NLRC4 / type: protein_or_peptide / ID: 1 / Name.synonym: IPAF / Number of copies: 11 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 100 KDa / Theoretical: 100 KDa |
| Recombinant expression | Organism:  |
-Macromolecule #2: PrgJ
| Macromolecule | Name: PrgJ / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) |
| Recombinant expression | Organism:  |
-Macromolecule #3: NAIP2
| Macromolecule | Name: NAIP2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 10 mM Tris-HCl pH 8.0, 100 mM NaCl and 10 mM DTT |
| Grid | Details: 200 mesh copper grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 110 K / Instrument: FEI VITROBOT MARK IV / Method: Blot for 1 second |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 22,500 times magnification |
| Date | May 26, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 (4k x 4k) / Number real images: 2039 / Average electron dose: 46 e/Å2 Details: Every image is the average of 32 frames recorded by the direct electron detector |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 37879 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)