+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30989 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of PEP bound Enolase from Mycobacterium tuberculosis | |||||||||
 Map data Map data | This is the final combined B factor sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | METAL BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationphosphopyruvate hydratase / phosphopyruvate hydratase complex / phosphopyruvate hydratase activity / glycolytic process / cell surface / magnesium ion binding / extracellular region Similarity search - Function | |||||||||
| Biological species |  | |||||||||
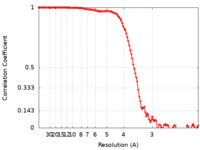
| Method | single particle reconstruction / cryo EM / Resolution: 3.23 Å | |||||||||
 Authors Authors | Bose S / Vinothkumar KR | |||||||||
| Funding support |  India, 2 items India, 2 items
| |||||||||
 Citation Citation |  Journal: IUCrJ / Year: 2023 Journal: IUCrJ / Year: 2023Title: Structural snapshots of Mycobacterium tuberculosis enolase reveal dual mode of 2PG binding and its implication in enzyme catalysis. Authors: Mohammed Ahmad / Bhavya Jha / Sucharita Bose / Satish Tiwari / Abhisek Dwivedy / Deepshikha Kar / Ravikant Pal / Richard Mariadasse / Tanya Parish / Jeyaraman Jeyakanthan / Kutti R ...Authors: Mohammed Ahmad / Bhavya Jha / Sucharita Bose / Satish Tiwari / Abhisek Dwivedy / Deepshikha Kar / Ravikant Pal / Richard Mariadasse / Tanya Parish / Jeyaraman Jeyakanthan / Kutti R Vinothkumar / Bichitra Kumar Biswal /   Abstract: Enolase, a ubiquitous enzyme, catalyzes the reversible conversion of 2-phosphoglycerate (2PG) to phosphoenolpyruvate (PEP) in the glycolytic pathway of organisms of all three domains of life. The ...Enolase, a ubiquitous enzyme, catalyzes the reversible conversion of 2-phosphoglycerate (2PG) to phosphoenolpyruvate (PEP) in the glycolytic pathway of organisms of all three domains of life. The underlying mechanism of the 2PG to PEP conversion has been studied in great detail in previous work, however that of the reverse reaction remains to be explored. Here we present structural snapshots of Mycobacterium tuberculosis (Mtb) enolase in apo, PEP-bound and two 2PG-bound forms as it catalyzes the conversion of PEP to 2PG. The two 2PG-bound complex structures differed in the conformation of the bound product (2PG) viz the widely reported canonical conformation and a novel binding pose, which we refer to here as the alternate conformation. Notably, we observed two major differences compared with the forward reaction: the presence of Mg is non-obligatory for the reaction and 2PG assumes an alternate conformation that is likely to facilitate its dissociation from the active site. Molecular dynamics studies and binding free energy calculations further substantiate that the alternate conformation of 2PG causes distortions in both metal ion coordination and hydrogen-bonding interactions, resulting in an increased flexibility of the active-site loops and aiding product release. Taken together, this study presents a probable mechanism involved in PEP to 2PG catalysis that is likely to be mediated by the conformational change of 2PG at the active site. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30989.map.gz emd_30989.map.gz | 116.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30989-v30.xml emd-30989-v30.xml emd-30989.xml emd-30989.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30989_fsc.xml emd_30989_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_30989.png emd_30989.png | 72.2 KB | ||
| Filedesc metadata |  emd-30989.cif.gz emd-30989.cif.gz | 6.4 KB | ||
| Others |  emd_30989_additional_1.map.gz emd_30989_additional_1.map.gz emd_30989_additional_2.map.gz emd_30989_additional_2.map.gz | 93.3 MB 93.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30989 http://ftp.pdbj.org/pub/emdb/structures/EMD-30989 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30989 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30989 | HTTPS FTP |
-Validation report
| Summary document |  emd_30989_validation.pdf.gz emd_30989_validation.pdf.gz | 607.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30989_full_validation.pdf.gz emd_30989_full_validation.pdf.gz | 607.2 KB | Display | |
| Data in XML |  emd_30989_validation.xml.gz emd_30989_validation.xml.gz | 12.1 KB | Display | |
| Data in CIF |  emd_30989_validation.cif.gz emd_30989_validation.cif.gz | 16.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30989 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30989 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30989 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30989 | HTTPS FTP |
-Related structure data
| Related structure data |  7e51MC  6l7dC  7ckpC  7clkC  7cllC  7dlrC  7e4fC  7e4xC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30989.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30989.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the final combined B factor sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: This is one of the unsharpened half maps
| File | emd_30989_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is one of the unsharpened half maps | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: This is the other unsharpened half map
| File | emd_30989_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the other unsharpened half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Octameric Enolase enzyme
| Entire | Name: Octameric Enolase enzyme |
|---|---|
| Components |
|
-Supramolecule #1: Octameric Enolase enzyme
| Supramolecule | Name: Octameric Enolase enzyme / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 370 KDa |
-Macromolecule #1: Enolase
| Macromolecule | Name: Enolase / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO / EC number: phosphopyruvate hydratase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.962355 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
| Sequence | String: MHHHHHHMPI IEQVRAREIL DSRGNPTVEV EVALIDGTFA RAAVPSGAST GEHEAVELRD GGDRYGGKGV QKAVQAVLDE IGPAVIGLN ADDQRLVDQA LVDLDGTPDK SRLGGNAILG VSLAVAKAAA DSAELPLFRY VGGPNAHILP VPMMNILNGG A HADTAVDI ...String: MHHHHHHMPI IEQVRAREIL DSRGNPTVEV EVALIDGTFA RAAVPSGAST GEHEAVELRD GGDRYGGKGV QKAVQAVLDE IGPAVIGLN ADDQRLVDQA LVDLDGTPDK SRLGGNAILG VSLAVAKAAA DSAELPLFRY VGGPNAHILP VPMMNILNGG A HADTAVDI QEFMVAPIGA PSFVEALRWG AEVYHALKSV LKKEGLSTGL GDEGGFAPDV AGTTAALDLI SRAIESAGLR PG ADVALAL DAAATEFFTD GTGYVFEGTT RTADQMTEFY AGLLGAYPLV SIEDPLSEDD WDGWAALTAS IGDRVQIVGD DIF VTNPER LEEGIERGVA NALLVKVNQI GTLTETLDAV TLAHHGGYRT MISHRSGETE DTMIADLAVA IGSGQIKTGA PARS ERVAK YNQLLRIEEA LGDAARYAGD LAFPRFACET K UniProtKB: Enolase |
-Macromolecule #2: PHOSPHOENOLPYRUVATE
| Macromolecule | Name: PHOSPHOENOLPYRUVATE / type: ligand / ID: 2 / Number of copies: 8 / Formula: PEP |
|---|---|
| Molecular weight | Theoretical: 168.042 Da |
| Chemical component information |  ChemComp-PEP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 16 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| |||||||||||||||
| Grid | Model: Quantifoil / Material: GOLD / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 289 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Average electron dose: 27.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 130841 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.813 µm / Nominal defocus min: 0.931 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z
Z Y
Y X
X