[English] 日本語
 Yorodumi
Yorodumi- EMDB-30944: Cryo-EM structure of hybrid respiratory supercomplex consisting o... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30944 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of hybrid respiratory supercomplex consisting of Mycobacterium tuberculosis complexIII and Mycobacterium smegmatis complexIV in the presence of Q203 | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationaerobic electron transport chain /  cytochrome-c oxidase / quinol-cytochrome-c reductase / cytochrome-c oxidase / quinol-cytochrome-c reductase /  ubiquinol-cytochrome-c reductase activity / ubiquinol-cytochrome-c reductase activity /  cytochrome-c oxidase activity / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen / cytochrome-c oxidase activity / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen /  respirasome / respiratory electron transport chain / peptidoglycan-based cell wall / respirasome / respiratory electron transport chain / peptidoglycan-based cell wall /  monooxygenase activity ...aerobic electron transport chain / monooxygenase activity ...aerobic electron transport chain /  cytochrome-c oxidase / quinol-cytochrome-c reductase / cytochrome-c oxidase / quinol-cytochrome-c reductase /  ubiquinol-cytochrome-c reductase activity / ubiquinol-cytochrome-c reductase activity /  cytochrome-c oxidase activity / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen / cytochrome-c oxidase activity / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen /  respirasome / respiratory electron transport chain / peptidoglycan-based cell wall / respirasome / respiratory electron transport chain / peptidoglycan-based cell wall /  monooxygenase activity / 2 iron, 2 sulfur cluster binding / monooxygenase activity / 2 iron, 2 sulfur cluster binding /  oxidoreductase activity / iron ion binding / copper ion binding / oxidoreductase activity / iron ion binding / copper ion binding /  heme binding / heme binding /  membrane / membrane /  metal ion binding / metal ion binding /  plasma membrane plasma membraneSimilarity search - Function | ||||||||||||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 51 (bacteria) / Mycolicibacterium smegmatis MC2 51 (bacteria) /  Mycobacterium smegmatis MC2 51 (bacteria) / Mycobacterium smegmatis MC2 51 (bacteria) /   Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) | ||||||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.67 Å cryo EM / Resolution: 2.67 Å | ||||||||||||||||||
 Authors Authors | Zhou S / Wang W / Gao Y / Gong H / Rao Z | ||||||||||||||||||
| Funding support |  China, 5 items China, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: Structure of cytochrome in complex with Q203 and TB47, two anti-TB drug candidates. Authors: Shan Zhou / Weiwei Wang / Xiaoting Zhou / Yuying Zhang / Yuezheng Lai / Yanting Tang / Jinxu Xu / Dongmei Li / Jianping Lin / Xiaolin Yang / Ting Ran / Hongming Chen / Luke W Guddat / Quan ...Authors: Shan Zhou / Weiwei Wang / Xiaoting Zhou / Yuying Zhang / Yuezheng Lai / Yanting Tang / Jinxu Xu / Dongmei Li / Jianping Lin / Xiaolin Yang / Ting Ran / Hongming Chen / Luke W Guddat / Quan Wang / Yan Gao / Zihe Rao / Hongri Gong /   Abstract: Pathogenic mycobacteria pose a sustained threat to global human health. Recently, cytochrome complexes have gained interest as targets for antibiotic drug development. However, there is currently no ...Pathogenic mycobacteria pose a sustained threat to global human health. Recently, cytochrome complexes have gained interest as targets for antibiotic drug development. However, there is currently no structural information for the cytochrome complex from these pathogenic mycobacteria. Here, we report the structures of cytochrome alone (2.68 Å resolution) and in complex with clinical drug candidates Q203 (2.67 Å resolution) and TB47 (2.93 Å resolution) determined by single-particle cryo-electron microscopy. cytochrome forms a dimeric assembly with endogenous menaquinone/menaquinol bound at the quinone/quinol-binding pockets. We observe Q203 and TB47 bound at the quinol-binding site and stabilized by hydrogen bonds with the side chains of Thr and Glu, residues that are conserved across pathogenic mycobacteria. These high-resolution images provide a basis for the design of new mycobacterial cytochrome inhibitors that could be developed into broad-spectrum drugs to treat mycobacterial infections. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30944.map.gz emd_30944.map.gz | 483.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30944-v30.xml emd-30944-v30.xml emd-30944.xml emd-30944.xml | 25.9 KB 25.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30944.png emd_30944.png | 166.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30944 http://ftp.pdbj.org/pub/emdb/structures/EMD-30944 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30944 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30944 | HTTPS FTP |
-Related structure data
| Related structure data |  7e1wMC  7e1vC  7e1xC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30944.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30944.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : hybrid respiratory supercomplex consisting of Mycobacterium tuber...
+Supramolecule #1: hybrid respiratory supercomplex consisting of Mycobacterium tuber...
+Macromolecule #1: Cytochrome c oxidase subunit 2
+Macromolecule #2: Cytochrome c oxidase subunit 1
+Macromolecule #3: Cytochrome c oxidase subunit 3
+Macromolecule #4: Cytochrome c oxidase polypeptide 4
+Macromolecule #5: Cytochrome c oxidase subunit CtaJ
+Macromolecule #6: Uncharacterized protein MSMEG_4692/MSMEI_4575
+Macromolecule #7: Prokaryotic respiratory supercomplex associate factor 1 PRSAF1
+Macromolecule #8: Cytochrome bc1 complex cytochrome b subunit
+Macromolecule #9: Cytochrome bc1 complex Rieske iron-sulfur subunit
+Macromolecule #10: Cytochrome bc1 complex cytochrome c subunit
+Macromolecule #11: COPPER (II) ION
+Macromolecule #12: CARDIOLIPIN
+Macromolecule #13: PALMITIC ACID
+Macromolecule #14: HEME-A
+Macromolecule #15: (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)...
+Macromolecule #16: PROTOPORPHYRIN IX CONTAINING FE
+Macromolecule #17: MENAQUINONE-9
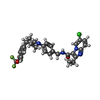
+Macromolecule #18: 6-chloranyl-2-ethyl-N-[[4-[4-[4-(trifluoromethyloxy)phenyl]piperi...
+Macromolecule #19: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #20: (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3...
+Macromolecule #21: HEME C
+Macromolecule #22: water
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 1.8 µm / Calibrated defocus min: 1.2 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
|---|---|
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.67 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 106770 |
 Movie
Movie Controller
Controller