[English] 日本語
 Yorodumi
Yorodumi- EMDB-30936: Molecular insights into ago-allosteric modulation of the human gl... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30936 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationglucagon-like peptide 1 receptor activity / Olfactory Signaling Pathway / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / glucagon receptor activity / hormone secretion / positive regulation of blood pressure / Activation of the phototransduction cascade / post-translational protein targeting to membrane, translocation / Activation of G protein gated Potassium channels ...glucagon-like peptide 1 receptor activity / Olfactory Signaling Pathway / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / glucagon receptor activity / hormone secretion / positive regulation of blood pressure / Activation of the phototransduction cascade / post-translational protein targeting to membrane, translocation / Activation of G protein gated Potassium channels / G-protein activation / G beta:gamma signalling through PI3Kgamma / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through PLC beta / ADP signalling through P2Y purinoceptor 1 / Thromboxane signalling through TP receptor / Presynaptic function of Kainate receptors / G beta:gamma signalling through CDC42 / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Glucagon-type ligand receptors / G alpha (12/13) signalling events / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Adrenaline,noradrenaline inhibits insulin secretion / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Thrombin signalling through proteinase activated receptors (PARs) / Ca2+ pathway / Extra-nuclear estrogen signaling / G alpha (z) signalling events / G alpha (s) signalling events / G alpha (q) signalling events / regulation of heart contraction / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (i) signalling events / response to psychosocial stress / Vasopressin regulates renal water homeostasis via Aquaporins / peptide hormone binding / activation of adenylate cyclase activity / cAMP-mediated signaling / negative regulation of blood pressure / adenylate cyclase-activating G protein-coupled receptor signaling pathway / photoreceptor disc membrane / Glucagon-type ligand receptors / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / cellular response to catecholamine stimulus / adenylate cyclase-activating dopamine receptor signaling pathway / cellular response to prostaglandin E stimulus / transmembrane signaling receptor activity / heterotrimeric G-protein complex / signaling receptor complex adaptor activity / positive regulation of cytosolic calcium ion concentration / G alpha (s) signalling events / learning or memory / cell surface receptor signaling pathway / G protein-coupled receptor signaling pathway / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
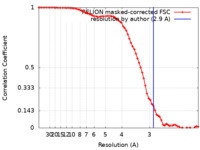
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Cong ZT / Chen LN / Ma HL / Zhou QT / Yang DH / Xu HE / Zhang Y / Wang MW | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor. Authors: Zhaotong Cong / Li-Nan Chen / Honglei Ma / Qingtong Zhou / Xinyu Zou / Chenyu Ye / Antao Dai / Qing Liu / Wei Huang / Xianqiang Sun / Xi Wang / Peiyu Xu / Lihua Zhao / Tian Xia / Wenge Zhong ...Authors: Zhaotong Cong / Li-Nan Chen / Honglei Ma / Qingtong Zhou / Xinyu Zou / Chenyu Ye / Antao Dai / Qing Liu / Wei Huang / Xianqiang Sun / Xi Wang / Peiyu Xu / Lihua Zhao / Tian Xia / Wenge Zhong / Dehua Yang / H Eric Xu / Yan Zhang / Ming-Wei Wang /  Abstract: The glucagon-like peptide-1 (GLP-1) receptor is a validated drug target for metabolic disorders. Ago-allosteric modulators are capable of acting both as agonists on their own and as efficacy ...The glucagon-like peptide-1 (GLP-1) receptor is a validated drug target for metabolic disorders. Ago-allosteric modulators are capable of acting both as agonists on their own and as efficacy enhancers of orthosteric ligands. However, the molecular details of ago-allosterism remain elusive. Here, we report three cryo-electron microscopy structures of GLP-1R bound to (i) compound 2 (an ago-allosteric modulator); (ii) compound 2 and GLP-1; and (iii) compound 2 and LY3502970 (a small molecule agonist), all in complex with heterotrimeric G. The structures reveal that compound 2 is covalently bonded to C347 at the cytoplasmic end of TM6 and triggers its outward movement in cooperation with the ECD whose N terminus penetrates into the GLP-1 binding site. This allows compound 2 to execute positive allosteric modulation through enhancement of both agonist binding and G protein coupling. Our findings offer insights into the structural basis of ago-allosterism at GLP-1R and may aid the design of better therapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30936.map.gz emd_30936.map.gz | 28.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30936-v30.xml emd-30936-v30.xml emd-30936.xml emd-30936.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30936_fsc.xml emd_30936_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_30936.png emd_30936.png | 22.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30936 http://ftp.pdbj.org/pub/emdb/structures/EMD-30936 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30936 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30936 | HTTPS FTP |
-Validation report
| Summary document |  emd_30936_validation.pdf.gz emd_30936_validation.pdf.gz | 384.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30936_full_validation.pdf.gz emd_30936_full_validation.pdf.gz | 384 KB | Display | |
| Data in XML |  emd_30936_validation.xml.gz emd_30936_validation.xml.gz | 9.7 KB | Display | |
| Data in CIF |  emd_30936_validation.cif.gz emd_30936_validation.cif.gz | 12.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30936 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30936 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30936 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30936 | HTTPS FTP |
-Related structure data
| Related structure data |  7e14MC  7duqC  7durC  7evmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30936.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30936.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : compound2_GLP1R_OWL833_Gs-complex
| Entire | Name: compound2_GLP1R_OWL833_Gs-complex |
|---|---|
| Components |
|
-Supramolecule #1: compound2_GLP1R_OWL833_Gs-complex
| Supramolecule | Name: compound2_GLP1R_OWL833_Gs-complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism: Insect cell expression vector pTIE1 (others) |
-Macromolecule #1: compound2_GLP1R_OWL833_Gs-complex
| Macromolecule | Name: compound2_GLP1R_OWL833_Gs-complex / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RPQGATVSLW ETVQKWREYR RQCQRSLTED PPPATDLFCN RTFDEYACWP DGEPGSFVNV SCPWYLPWAS SVPQGHVYRF CTAEGLWLQK DNSSLPWRDL SECEESKRGE RSSPEEQLLF LYIIYTVGYA LSFSALVIAS AILLGFRHLH CTRNYIHLNL FASFILRALS ...String: RPQGATVSLW ETVQKWREYR RQCQRSLTED PPPATDLFCN RTFDEYACWP DGEPGSFVNV SCPWYLPWAS SVPQGHVYRF CTAEGLWLQK DNSSLPWRDL SECEESKRGE RSSPEEQLLF LYIIYTVGYA LSFSALVIAS AILLGFRHLH CTRNYIHLNL FASFILRALS VFIKDAALKW MYSTAAQQHQ WDGLLSYQDS LSCRLVFLLM QYCVAANYYW LLVEGVYLYT LLAFSVLSEQ WIFRLYVSIG WGVPLLFVVP WGIVKYLYED EGCWTRNSNM NYWLIIRLPI LFAIGVNFLI FVRVICIVVS KLKANLMCKT DIKCRLAKST LTLIPLLGTH EVIFAFVMDE HARGTLRFIK LFTELSFTSF QGLMVAILYC FVNNEVQLEF RKSWERWRLE HLHIQRDSSM KPLKCPTSSL SSGATAGSSM YTATCQASCS MGCLGNSKT EDQRNEEKAQ REANKKIEKQ LQKDKQVYRA THRLLLLGAD NSGKSTIVKQ MRIYHVNSGI FETKFQVDKV NFHMFDVGAQ RDERRKWIQC FNDVTAIIFV VDSSDYNRLQ AALKLFDSIW NNKWLRDTSV ILFLNKQDLL AEKVLAGKSK IEDYFPEFAR YTTPEDATPE PGEDPRVTRA KYFIRDEFLR ISTASGDGRH YCYPHFTCSV DTENIRRVFN DCRDIIQRMH LRQYELL MG SLLQSELDQL RQEAEQLKNQ IRDARKACAD ATLSQITNNI DPVGRIQMRT RRTLRGHLAK IYAMHWGTDS RLLVSASQDG KLIIWDSYTT NKVHAIPLRS SWVMTCAYAP SGNYVACGGL DNICSIYNLK TREGNVRVSR ELAGHTGYLS CCRFLDDNQI VTSSGDTTCA LWDIETGQQT TTFTGHTGDV MSLSLAPDTR LFVSGACDAS AKLWDVREGM CRQTFTGHES DINAICFFPN GNAFATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGVTDDGMAV ATGSWDSFLK IWN MASNNT ASIAQARKLV EQLKMEANID RIKVSKAAAD LMAYCEAHAK EDPLLTPVPA SENPFREKKF FCAIL QVQL QESGGGLVQP GGSLRLSCAA SGFTFSNYKM NWVRQAPGKG LEWVSDISQS GASISYTGSV KGRFTISRDN AKNTLYLQMN SLKPEDTAVY YCARCPAPFT RDCFDVTSTT YAYRGQGTQV TVSS |
| Recombinant expression | Organism: Insect cell expression vector pTIE1 (others) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 101.325 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)