[English] 日本語
 Yorodumi
Yorodumi- EMDB-30657: Cryo-EM structure of a heme-copper terminal oxidase dimer provide... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30657 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a heme-copper terminal oxidase dimer provides insights into its catalytic mechanism | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | electron cryo-microscopy / heme-copper oxidase / cytochrome c oxidase dimer / Aquifex aeolicus / naphthoquinone / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationcell envelope / cytochrome-c oxidase activity / aerobic respiration / copper ion binding / heme binding / membrane Similarity search - Function | |||||||||
| Biological species |   Aquifex aeolicus VF5 (bacteria) / Aquifex aeolicus VF5 (bacteria) /   Aquifex aeolicus (strain VF5) (bacteria) / Aquifex aeolicus (strain VF5) (bacteria) /   Aquifex aeolicus (bacteria) Aquifex aeolicus (bacteria) | |||||||||
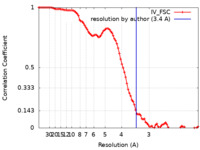
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Fei S / Hartmut M | |||||||||
 Citation Citation |  Journal: Angew Chem Int Ed Engl / Year: 2021 Journal: Angew Chem Int Ed Engl / Year: 2021Title: The Unusual Homodimer of a Heme-Copper Terminal Oxidase Allows Itself to Utilize Two Electron Donors. Authors: Guoliang Zhu / Hui Zeng / Shuangbo Zhang / Jana Juli / Linhua Tai / Danyang Zhang / Xiaoyun Pang / Yan Zhang / Sin Man Lam / Yun Zhu / Guohong Peng / Hartmut Michel / Fei Sun /   Abstract: The heme-copper oxidase superfamily comprises cytochrome c and ubiquinol oxidases. These enzymes catalyze the transfer of electrons from different electron donors onto molecular oxygen. A B-family ...The heme-copper oxidase superfamily comprises cytochrome c and ubiquinol oxidases. These enzymes catalyze the transfer of electrons from different electron donors onto molecular oxygen. A B-family cytochrome c oxidase from the hyperthermophilic bacterium Aquifex aeolicus was discovered previously to be able to use both cytochrome c and naphthoquinol as electron donors. Its molecular mechanism as well as the evolutionary significance are yet unknown. Here we solved its 3.4 Å resolution electron cryo-microscopic structure and discovered a novel dimeric structure mediated by subunit I (CoxA2) that would be essential for naphthoquinol binding and oxidation. The unique structural features in both proton and oxygen pathways suggest an evolutionary adaptation of this oxidase to its hyperthermophilic environment. Our results add a new conceptual understanding of structural variation of cytochrome c oxidases in different species. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30657.map.gz emd_30657.map.gz | 5.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30657-v30.xml emd-30657-v30.xml emd-30657.xml emd-30657.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30657_fsc.xml emd_30657_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_30657.png emd_30657.png | 133.4 KB | ||
| Filedesc metadata |  emd-30657.cif.gz emd-30657.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30657 http://ftp.pdbj.org/pub/emdb/structures/EMD-30657 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30657 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30657 | HTTPS FTP |
-Validation report
| Summary document |  emd_30657_validation.pdf.gz emd_30657_validation.pdf.gz | 400.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30657_full_validation.pdf.gz emd_30657_full_validation.pdf.gz | 400.4 KB | Display | |
| Data in XML |  emd_30657_validation.xml.gz emd_30657_validation.xml.gz | 10.9 KB | Display | |
| Data in CIF |  emd_30657_validation.cif.gz emd_30657_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30657 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30657 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30657 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30657 | HTTPS FTP |
-Related structure data
| Related structure data |  7degMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30657.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30657.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
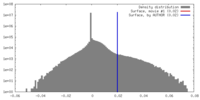
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : cytochrome c oxidase (respiratory complex IV)
+Supramolecule #1: cytochrome c oxidase (respiratory complex IV)
+Macromolecule #1: Cytochrome c oxidase subunit I
+Macromolecule #2: Cytochrome oxidase subunit IIa
+Macromolecule #3: Cytochrome oxidase subunit II
+Macromolecule #4: HEME-AS
+Macromolecule #5: PROTOPORPHYRIN IX CONTAINING FE
+Macromolecule #6: COPPER (II) ION
+Macromolecule #7: 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-h...
+Macromolecule #8: (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-...
+Macromolecule #9: 1,2-Distearoyl-sn-glycerophosphoethanolamine
+Macromolecule #10: DINUCLEAR COPPER ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)