+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | PP5 bound to Hsp90:Cdc37:CRaf complex | |||||||||||||||
 Map data Map data | PP5 bound to Hsp90:Cdc37:CRaf complex. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
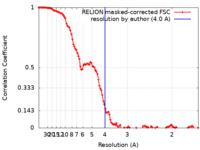
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||||||||
 Authors Authors | Jaime-Garza M / Nowotny CA / Coutandin D / Wang F / Tabios M / Agard DA | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Hsp90 provides a platform for kinase dephosphorylation by PP5. Authors: Maru Jaime-Garza / Carlos A Nowotny / Daniel Coutandin / Feng Wang / Mariano Tabios / David A Agard /  Abstract: The Hsp90 molecular chaperone collaborates with the phosphorylated Cdc37 cochaperone for the folding and activation of its many client kinases. As with many kinases, the Hsp90 client kinase CRaf is ...The Hsp90 molecular chaperone collaborates with the phosphorylated Cdc37 cochaperone for the folding and activation of its many client kinases. As with many kinases, the Hsp90 client kinase CRaf is activated by phosphorylation at specific regulatory sites. The cochaperone phosphatase PP5 dephosphorylates CRaf and Cdc37 in an Hsp90-dependent manner. Although dephosphorylating Cdc37 has been proposed as a mechanism for releasing Hsp90-bound kinases, here we show that Hsp90 bound kinases sterically inhibit Cdc37 dephosphorylation indicating kinase release must occur before Cdc37 dephosphorylation. Our cryo-EM structure of PP5 in complex with Hsp90:Cdc37:CRaf reveals how Hsp90 both activates PP5 and scaffolds its association with the bound CRaf to dephosphorylate phosphorylation sites neighboring the kinase domain. Thus, we directly show how Hsp90's role in maintaining protein homeostasis goes beyond folding and activation to include post translationally modifying its client kinases. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29973.map.gz emd_29973.map.gz | 98.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29973-v30.xml emd-29973-v30.xml emd-29973.xml emd-29973.xml | 25.8 KB 25.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29973_fsc.xml emd_29973_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_29973.png emd_29973.png | 88.7 KB | ||
| Others |  emd_29973_additional_1.map.gz emd_29973_additional_1.map.gz emd_29973_half_map_1.map.gz emd_29973_half_map_1.map.gz emd_29973_half_map_2.map.gz emd_29973_half_map_2.map.gz | 116.9 MB 98.4 MB 98.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29973 http://ftp.pdbj.org/pub/emdb/structures/EMD-29973 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29973 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29973 | HTTPS FTP |
-Validation report
| Summary document |  emd_29973_validation.pdf.gz emd_29973_validation.pdf.gz | 944.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29973_full_validation.pdf.gz emd_29973_full_validation.pdf.gz | 943.7 KB | Display | |
| Data in XML |  emd_29973_validation.xml.gz emd_29973_validation.xml.gz | 18.5 KB | Display | |
| Data in CIF |  emd_29973_validation.cif.gz emd_29973_validation.cif.gz | 24.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29973 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29973 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29973 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29973 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29973.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29973.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PP5 bound to Hsp90:Cdc37:CRaf complex. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: PostProcessed, sharpened map.
| File | emd_29973_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PostProcessed, sharpened map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
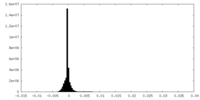
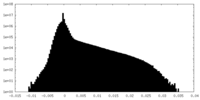
| Density Histograms |
-Half map: Half map before postprocessing.
| File | emd_29973_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map before postprocessing. | ||||||||||||
| Projections & Slices |
| ||||||||||||
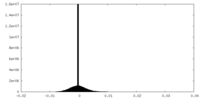
| Density Histograms |
-Half map: Half map before postprocessing.
| File | emd_29973_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map before postprocessing. | ||||||||||||
| Projections & Slices |
| ||||||||||||
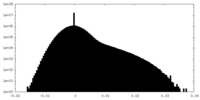
| Density Histograms |
- Sample components
Sample components
-Entire : Hsp90:Cdc37:CRaf complex bound to PP5.
| Entire | Name: Hsp90:Cdc37:CRaf complex bound to PP5. |
|---|---|
| Components |
|
-Supramolecule #1: Hsp90:Cdc37:CRaf complex bound to PP5.
| Supramolecule | Name: Hsp90:Cdc37:CRaf complex bound to PP5. / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all Details: Yeast purified Hsp90:Cdc37:CRaf complex incubated with e. Coli purified PP5 (mutant H304A). Sample then cross-linked with 0.05% glutaraldehyde for 15m at room temperature, and ran over S200 ...Details: Yeast purified Hsp90:Cdc37:CRaf complex incubated with e. Coli purified PP5 (mutant H304A). Sample then cross-linked with 0.05% glutaraldehyde for 15m at room temperature, and ran over S200 sizing column. Particles were then classified to yield a Hsp90:Cdc37:CRaf:PP5 complex. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 57 KDa |
-Supramolecule #2: Protein Phosphatase 5 (H304A)
| Supramolecule | Name: Protein Phosphatase 5 (H304A) / type: complex / ID: 2 / Chimera: Yes / Parent: 1 / Macromolecule list: #4 Details: E coli purified Protein Phosphatase 5, with inactivating H304A mutation. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Heat shock protein HSP 90-beta
| Macromolecule | Name: Heat shock protein HSP 90-beta / type: protein_or_peptide / ID: 1 Details: HRV 3C cleavage site, followed by a glycine linker and the Hsp90B sequence. Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: GPGMPEEVHH GEEEVETFAF QAEIAQLMSL IINTFYSNKE IFLRELISNA SDALDKIRYE SLTDPSKLDS GKELKIDIIP NPQERTLTLV DTGIGMTKAD LINNLGTIAK SGTKAFMEAL QAGADISMIG QFGVGFYSAY LVAEKVVVIT KHNDDEQYAW ESSAGGSFTV ...String: GPGMPEEVHH GEEEVETFAF QAEIAQLMSL IINTFYSNKE IFLRELISNA SDALDKIRYE SLTDPSKLDS GKELKIDIIP NPQERTLTLV DTGIGMTKAD LINNLGTIAK SGTKAFMEAL QAGADISMIG QFGVGFYSAY LVAEKVVVIT KHNDDEQYAW ESSAGGSFTV RADHGEPIGR GTKVILHLKE DQTEYLEERR VKEVVKKHSQ FIGYPITLYL EKEREKEISD DEAEEEKGEK EEEDKDDEEK PKIEDVGSDE EDDSGKDKKK KTKKIKEKYI DQEELNKTKP IWTRNPDDIT QEEYGEFYKS LTNDWEDHLA VKHFSVEGQL EFRALLFIPR RAPFDLFENK KKKNNIKLYV RRVFIMDSCD ELIPEYLNFI RGVVDSEDLP LNISREMLQQ SKILKVIRKN IVKKCLELFS ELAEDKENYK KFYEAFSKNL KLGIHEDSTN RRRLSELLRY HTSQSGDEMT SLSEYVSRMK ETQKSIYYIT GESKEQVANS AFVERVRKRG FEVVYMTEPI DEYCVQQLKE FDGKSLVSVT KEGLELPEDE EEKKKMEESK AKFENLCKLM KEILDKKVEK VTISNRLVSS PCCIVTSTYG WTANMERIMK AQALRDNSTM GYMMAKKHLE INPDHPIVET LRQKAEADKN DKAVKDLVVL LFETALLSSG FSLEDPQTHS NRIYRMIKLG LGIDEDEVAA EEPNAAVPDE IPPLEGDEDA SRMEEVD |
-Macromolecule #2: Hsp90 co-chaperone Cdc37
| Macromolecule | Name: Hsp90 co-chaperone Cdc37 / type: protein_or_peptide / ID: 2 Details: Cdc37 sequence followed by HRV 3C cleavage site (LEVLFQ) Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MVDYSVWDHI EVSDDEDETH PNIDTASLFR WRHQARVERM EQFQKEKEEL DRGCRECKRK VAECQRKLKE LEVAEGGKAE LERLQAEAQQ LRKEERSWEQ KLEEMRKKEK SMPWNVDTLS KDGFSKSMVN TKPEKTEEDS EEVREQKHKT FVEKYEKQIK HFGMLRRWDD ...String: MVDYSVWDHI EVSDDEDETH PNIDTASLFR WRHQARVERM EQFQKEKEEL DRGCRECKRK VAECQRKLKE LEVAEGGKAE LERLQAEAQQ LRKEERSWEQ KLEEMRKKEK SMPWNVDTLS KDGFSKSMVN TKPEKTEEDS EEVREQKHKT FVEKYEKQIK HFGMLRRWDD SQKYLSDNVH LVCEETANYL VIWCIDLEVE EKCALMEQVA HQTIVMQFIL ELAKSLKVDP RACFRQFFTK IKTADRQYME GFNDELEAFK ERVRGRAKLR IEKAMKEYEE EERKKRLGPG GLDPVEVYES LPEELQKCFD VKDVQMLQDA ISKMDPTDAK YHMQRCIDSG LWVPNSKASE AKEGEEAGPG DPLLEAVPKT GDEKDVSVLE VLFQ |
-Macromolecule #3: RAF proto-oncogene serine/threonine-protein kinase
| Macromolecule | Name: RAF proto-oncogene serine/threonine-protein kinase / type: protein_or_peptide / ID: 3 Details: Truncated CRaf (RAF-1) kinase domain followed by Strep Tag II sequence (WSHPQFEK) and HRV 3C cleavage site (LEVLFQ) Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: GGRDSSYYWE IEASEVMLST RIGSGSFGTV YKGKWHGDVA VKILKVVDPT PEQFQAFRNE VAVLRKTRHV NILLFMGYMT KDNLAIVTQW CEGSSLYKHL HVQETKFQMF QLIDIARQTA QGMDYLHAKN IIHRDMKSNN IFLHEGLTVK IGDFGLATVK SRWSGSQQVE ...String: GGRDSSYYWE IEASEVMLST RIGSGSFGTV YKGKWHGDVA VKILKVVDPT PEQFQAFRNE VAVLRKTRHV NILLFMGYMT KDNLAIVTQW CEGSSLYKHL HVQETKFQMF QLIDIARQTA QGMDYLHAKN IIHRDMKSNN IFLHEGLTVK IGDFGLATVK SRWSGSQQVE QPTGSVLWMA PEVIRMQDNN PFSFQSDVYS YGIVLYELMT GELPYSHINN RDQIIFMVGR GYASPDLSKL YKNCPKAMKR LVADCVKKVK EERPLFPQIL SSIELLQHSL PKINRLPESG WSHPQFEKLE VLFQ |
-Macromolecule #4: Serine/threonine-protein phosphatase 5
| Macromolecule | Name: Serine/threonine-protein phosphatase 5 / type: protein_or_peptide / ID: 4 Details: HRV 3C cleavage site followed by GS linker and PP5 sequence. Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: GPGSMAMAEG ERTECAEPPR DEPPADGALK RAEELKTQAN DYFKAKDYEN AIKFYSQAIE LNPSNAIYYG NRSLAYLRTE CYGYALGDAT RAIELDKKYI KGYYRRAASN MALGKFRAAL RDYETVVKVK PHDKDAKMKY QECNKIVKQK AFERAIAGDE HKRSVVDSLD ...String: GPGSMAMAEG ERTECAEPPR DEPPADGALK RAEELKTQAN DYFKAKDYEN AIKFYSQAIE LNPSNAIYYG NRSLAYLRTE CYGYALGDAT RAIELDKKYI KGYYRRAASN MALGKFRAAL RDYETVVKVK PHDKDAKMKY QECNKIVKQK AFERAIAGDE HKRSVVDSLD IESMTIEDEY SGPKLEDGKV TISFMKELMQ WYKDQKKLHR KCAYQILVQV KEVLSKLSTL VETTLKETEK ITVCGDTHGQ FYDLLNIFEL NGLPSETNPY IFNGDFVDRG SFSVEVILTL FGFKLLYPDH FHLLRGNAET DNMNQIYGFE GEVKAKYTAQ MYELFSEVFE WLPLAQCING KVLIMHGGLF SEDGVTLDDI RKIERNRQPP DSGPMCDLLW SDPQPQNGRS ISKRGVSCQF GPDVTKAFLE ENNLDYIIRS HEVKAEGYEV AHGGRCVTVF SAPNYCDQMG NKASYIHLQG SDLRPQFHQF TAVPHPNVKP MAYANTLLQL GMM |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Support film - Film thickness: 1.0 nm | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV Details: Sample Vol: 3UL Temp: 10C Humidity: 100% WAIT TIME: 30S BLOT TIME: 3S BLOT FORCE: -2. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 4160 / Average electron dose: 69.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)