+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Hsp90:Cdc37:CRaf complex | |||||||||||||||
 Map data Map data | This map was obtained through focused classification with subtraction of the Cdc37 Middle domain and CRaf N-lobe portion of this Hsp90:Cdc37:CRaf complex, and further refinement. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
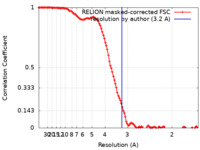
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||||||||
 Authors Authors | Jaime-Garza M / Nowotny CA / Coutandin D / Wang F / Tabios M / Agard DA | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Hsp90 provides a platform for kinase dephosphorylation by PP5. Authors: Maru Jaime-Garza / Carlos A Nowotny / Daniel Coutandin / Feng Wang / Mariano Tabios / David A Agard /  Abstract: The Hsp90 molecular chaperone collaborates with the phosphorylated Cdc37 cochaperone for the folding and activation of its many client kinases. As with many kinases, the Hsp90 client kinase CRaf is ...The Hsp90 molecular chaperone collaborates with the phosphorylated Cdc37 cochaperone for the folding and activation of its many client kinases. As with many kinases, the Hsp90 client kinase CRaf is activated by phosphorylation at specific regulatory sites. The cochaperone phosphatase PP5 dephosphorylates CRaf and Cdc37 in an Hsp90-dependent manner. Although dephosphorylating Cdc37 has been proposed as a mechanism for releasing Hsp90-bound kinases, here we show that Hsp90 bound kinases sterically inhibit Cdc37 dephosphorylation indicating kinase release must occur before Cdc37 dephosphorylation. Our cryo-EM structure of PP5 in complex with Hsp90:Cdc37:CRaf reveals how Hsp90 both activates PP5 and scaffolds its association with the bound CRaf to dephosphorylate phosphorylation sites neighboring the kinase domain. Thus, we directly show how Hsp90's role in maintaining protein homeostasis goes beyond folding and activation to include post translationally modifying its client kinases. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29949.map.gz emd_29949.map.gz | 98.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29949-v30.xml emd-29949-v30.xml emd-29949.xml emd-29949.xml | 23.3 KB 23.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29949_fsc.xml emd_29949_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_29949.png emd_29949.png | 96.3 KB | ||
| Others |  emd_29949_additional_1.map.gz emd_29949_additional_1.map.gz emd_29949_half_map_1.map.gz emd_29949_half_map_1.map.gz emd_29949_half_map_2.map.gz emd_29949_half_map_2.map.gz | 117.2 MB 98.7 MB 98.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29949 http://ftp.pdbj.org/pub/emdb/structures/EMD-29949 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29949 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29949 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29949.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29949.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This map was obtained through focused classification with subtraction of the Cdc37 Middle domain and CRaf N-lobe portion of this Hsp90:Cdc37:CRaf complex, and further refinement. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Postprocessed map.
| File | emd_29949_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map.
| File | emd_29949_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map.
| File | emd_29949_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Hsp90:Cdc37:CRaf complex
| Entire | Name: Hsp90:Cdc37:CRaf complex |
|---|---|
| Components |
|
-Supramolecule #1: Hsp90:Cdc37:CRaf complex
| Supramolecule | Name: Hsp90:Cdc37:CRaf complex / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all Details: Yeast purified Hsp90:Cdc37:CRaf complex incubated with e. Coli purified PP5 (mutant H304A). Sample then cross-linked with 0.05% glutaraldehyde for 15m at room temperature, and ran over S200 ...Details: Yeast purified Hsp90:Cdc37:CRaf complex incubated with e. Coli purified PP5 (mutant H304A). Sample then cross-linked with 0.05% glutaraldehyde for 15m at room temperature, and ran over S200 sizing column. Particles were then classified to yield a Hsp90:Cdc37:CRaf complex. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 57 KDa |
-Macromolecule #1: Heat shock protein HSP 90-beta
| Macromolecule | Name: Heat shock protein HSP 90-beta / type: protein_or_peptide / ID: 1 Details: Sequence starts with HRV 3C cleavage site, followed by human Hsp90B sequence. Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: GPGMPEEVHH GEEEVETFAF QAEIAQLMSL IINTFYSNKE IFLRELISNA SDALDKIRYE SLTDPSKLDS GKELKIDIIP NPQERTLTL VDTGIGMTKA DLINNLGTIA KSGTKAFMEA LQAGADISMI GQFGVGFYSA YLVAEKVVVI TKHNDDEQYA W ESSAGGSF ...String: GPGMPEEVHH GEEEVETFAF QAEIAQLMSL IINTFYSNKE IFLRELISNA SDALDKIRYE SLTDPSKLDS GKELKIDIIP NPQERTLTL VDTGIGMTKA DLINNLGTIA KSGTKAFMEA LQAGADISMI GQFGVGFYSA YLVAEKVVVI TKHNDDEQYA W ESSAGGSF TVRADHGEPI GRGTKVILHL KEDQTEYLEE RRVKEVVKKH SQFIGYPITL YLEKEREKEI SDDEAEEEKG EK EEEDKDD EEKPKIEDVG SDEEDDSGKD KKKKTKKIKE KYIDQEELNK TKPIWTRNPD DITQEEYGEF YKSLTNDWED HLA VKHFSV EGQLEFRALL FIPRRAPFDL FENKKKKNNI KLYVRRVFIM DSCDELIPEY LNFIRGVVDS EDLPLNISRE MLQQ SKILK VIRKNIVKKC LELFSELAED KENYKKFYEA FSKNLKLGIH EDSTNRRRLS ELLRYHTSQS GDEMTSLSEY VSRMK ETQK SIYYITGESK EQVANSAFVE RVRKRGFEVV YMTEPIDEYC VQQLKEFDGK SLVSVTKEGL ELPEDEEEKK KMEESK AKF ENLCKLMKEI LDKKVEKVTI SNRLVSSPCC IVTSTYGWTA NMERIMKAQA LRDNSTMGYM MAKKHLEINP DHPIVET LR QKAEADKNDK AVKDLVVLLF ETALLSSGFS LEDPQTHSNR IYRMIKLGLG IDEDEVAAEE PNAAVPDEIP PLEGDEDA S RMEEVD |
-Macromolecule #2: Hsp90 co-chaperone Cdc37
| Macromolecule | Name: Hsp90 co-chaperone Cdc37 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MVDYSVWDHI EV(SEP)DDEDETH PNIDTASLFR WRHQARVERM EQFQKEKEEL DRGCRECKRK VAECQRKLKE LEVAEG GKA ELERLQAEAQ QLRKEERSWE QKLEEMRKKE KSMPWNVDTL SKDGFSKSMV NTKPEKTEED SEEVREQKHK TFVEKYE KQ IKHFGMLRRW ...String: MVDYSVWDHI EV(SEP)DDEDETH PNIDTASLFR WRHQARVERM EQFQKEKEEL DRGCRECKRK VAECQRKLKE LEVAEG GKA ELERLQAEAQ QLRKEERSWE QKLEEMRKKE KSMPWNVDTL SKDGFSKSMV NTKPEKTEED SEEVREQKHK TFVEKYE KQ IKHFGMLRRW DDSQKYLSDN VHLVCEETAN YLVIWCIDLE VEEKCALMEQ VAHQTIVMQF ILELAKSLKV DPRACFRQ F FTKIKTADRQ YMEGFNDELE AFKERVRGRA KLRIEKAMKE YEEEERKKRL GPGGLDPVEV YESLPEELQK CFDVKDVQM LQDAISKMDP TDAKYHMQRC IDSGLWVPNS KASEAKEGEE AGPGDPLLEA VPKTGDEKDV SVLEVLFQ |
-Macromolecule #3: RAF proto-oncogene serine/threonine-protein kinase
| Macromolecule | Name: RAF proto-oncogene serine/threonine-protein kinase / type: protein_or_peptide / ID: 3 Details: Raf1 kinase sequence residues 336-618, followed by linker, Strep Tag II (WSHPQFEK) and HRV 3C site (LEVLFQ). Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: NILLFMGYMT KDNLAIVTQW CEGSSLYKHL HVQETKFQMF QLIDIARQTA QGMDYLHAKN IIHRDMKSNN IFLHEGLTVK IGDFGLATV KSRWSGSQQV EQPTGSVLWM APEVIRMQDN NPFSFQSDVY SYGIVLYELM TGELPYSHIN NRDQIIFMVG R GYASPDLS ...String: NILLFMGYMT KDNLAIVTQW CEGSSLYKHL HVQETKFQMF QLIDIARQTA QGMDYLHAKN IIHRDMKSNN IFLHEGLTVK IGDFGLATV KSRWSGSQQV EQPTGSVLWM APEVIRMQDN NPFSFQSDVY SYGIVLYELM TGELPYSHIN NRDQIIFMVG R GYASPDLS KLYKNCPKAM KRLVADCVKK VKEERPLFPQ ILSSIELLQ |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Support film - Film thickness: 1.0 nm | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV Details: Sample Vol: 3UL Temp: 10C Humidity: 100% WAIT TIME: 30S BLOT TIME: 3S BLOT FORCE: -2. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 4160 / Average electron dose: 69.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)