[English] 日本語
 Yorodumi
Yorodumi- EMDB-29744: local refinement of the primase/DNA/primer region in mutant T4 pr... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | local refinement of the primase/DNA/primer region in mutant T4 primosome | |||||||||
 Map data Map data | focus refinement of the primase/DNA-RNA region | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | phage / complex / helicase / REPLICATION | |||||||||
| Biological species |  Escherichia phage T4 (virus) Escherichia phage T4 (virus) | |||||||||
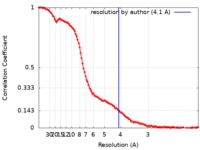
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Feng X / Li H | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis of the T4 bacteriophage primosome assembly and primer synthesis. Authors: Xiang Feng / Michelle M Spiering / Ruda de Luna Almeida Santos / Stephen J Benkovic / Huilin Li /  Abstract: The T4 bacteriophage gp41 helicase and gp61 primase assemble into a primosome to couple DNA unwinding with RNA primer synthesis for DNA replication. How the primosome is assembled and how the primer ...The T4 bacteriophage gp41 helicase and gp61 primase assemble into a primosome to couple DNA unwinding with RNA primer synthesis for DNA replication. How the primosome is assembled and how the primer length is defined are unclear. Here we report a series of cryo-EM structures of T4 primosome assembly intermediates. We show that gp41 alone is an open spiral, and ssDNA binding triggers a large-scale scissor-like conformational change that drives the ring closure and activates the helicase. Helicase activation exposes a cryptic hydrophobic surface to recruit the gp61 primase. The primase binds the helicase in a bipartite mode in which the N-terminal Zn-binding domain and the C-terminal RNA polymerase domain each contain a helicase-interacting motif that bind to separate gp41 N-terminal hairpin dimers, leading to the assembly of one primase on the helicase hexamer. Our study reveals the T4 primosome assembly process and sheds light on the RNA primer synthesis mechanism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29744.map.gz emd_29744.map.gz | 63.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29744-v30.xml emd-29744-v30.xml emd-29744.xml emd-29744.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29744_fsc.xml emd_29744_fsc.xml | 14.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_29744.png emd_29744.png | 44.1 KB | ||
| Others |  emd_29744_half_map_1.map.gz emd_29744_half_map_1.map.gz emd_29744_half_map_2.map.gz emd_29744_half_map_2.map.gz | 115.8 MB 115.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29744 http://ftp.pdbj.org/pub/emdb/structures/EMD-29744 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29744 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29744 | HTTPS FTP |
-Validation report
| Summary document |  emd_29744_validation.pdf.gz emd_29744_validation.pdf.gz | 725.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29744_full_validation.pdf.gz emd_29744_full_validation.pdf.gz | 725.4 KB | Display | |
| Data in XML |  emd_29744_validation.xml.gz emd_29744_validation.xml.gz | 18.1 KB | Display | |
| Data in CIF |  emd_29744_validation.cif.gz emd_29744_validation.cif.gz | 23.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29744 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29744 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29744 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29744 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29744.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29744.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | focus refinement of the primase/DNA-RNA region | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.029 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_29744_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_29744_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of mutated T4 bacteriophage helicase gp41 with ssDNA/RNA ...
| Entire | Name: Complex of mutated T4 bacteriophage helicase gp41 with ssDNA/RNA hybrid |
|---|---|
| Components |
|
-Supramolecule #1: Complex of mutated T4 bacteriophage helicase gp41 with ssDNA/RNA ...
| Supramolecule | Name: Complex of mutated T4 bacteriophage helicase gp41 with ssDNA/RNA hybrid type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Escherichia phage T4 (virus) Escherichia phage T4 (virus) |
-Macromolecule #1: Bacteriophage T4 DNA primase
| Macromolecule | Name: Bacteriophage T4 DNA primase / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO EC number: Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases |
|---|---|
| Source (natural) | Organism:  Escherichia phage T4 (virus) Escherichia phage T4 (virus) |
| Recombinant expression | Organism:  |
| Sequence | String: MSSIPWIDNE FAYRALAHLP KFTQVNNSST FKLRFRCPVC GDSKTDQNKA RGWYYGDNNE GNIHCYNCN YHAPIGIYLK EFEPDLYREY IFEIRKEKGK SRPIEKPKEL PKQPEKKIIK S LPSCVRLD KLAEDHPIIK YVKARCIPKD KWKYLWFTTE WPKLVNSIAP ...String: MSSIPWIDNE FAYRALAHLP KFTQVNNSST FKLRFRCPVC GDSKTDQNKA RGWYYGDNNE GNIHCYNCN YHAPIGIYLK EFEPDLYREY IFEIRKEKGK SRPIEKPKEL PKQPEKKIIK S LPSCVRLD KLAEDHPIIK YVKARCIPKD KWKYLWFTTE WPKLVNSIAP GTYKKEISEP RL VIPIYNA NGKAESFQGR ALKKDAPQKY ITIEAYPEAT KIYGVERVKD GDVYVLEGPI DSL FIENGI AITGGQLDLE VVPFKDRRVW VLDNEPRHPD TIKRMTKLVD AGERVMFWDK SPWK SKDVN DMIRKEGATP EQIMEYMKNN IAQGLMAKMR LSKYAKI |
-Macromolecule #2: DNA (70-mer)
| Macromolecule | Name: DNA (70-mer) / type: dna / ID: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Escherichia phage T4 (virus) Escherichia phage T4 (virus) |
| Sequence | String: (DG)(DA)(DA)(DT)(DG)(DA)(DG)(DG)(DA)(DG) (DT)(DA)(DG)(DT)(DA)(DG)(DT)(DG)(DA)(DA) (DT)(DG)(DT)(DA)(DG)(DT)(DG)(DA)(DG) (DG)(DT)(DA)(DA)(DT)(DA)(DT)(DC)(DG)(DG) (DC) (DT)(DG)(DG)(DT)(DC)(DT) ...String: (DG)(DA)(DA)(DT)(DG)(DA)(DG)(DG)(DA)(DG) (DT)(DA)(DG)(DT)(DA)(DG)(DT)(DG)(DA)(DA) (DT)(DG)(DT)(DA)(DG)(DT)(DG)(DA)(DG) (DG)(DT)(DA)(DA)(DT)(DA)(DT)(DC)(DG)(DG) (DC) (DT)(DG)(DG)(DT)(DC)(DT)(DG)(DG) (DT)(DC)(DT)(DG)(DT)(DG)(DC)(DC)(DA)(DA) (DG)(DT) (DT)(DG)(DC)(DT)(DG)(DC)(DA) (DA)(DA)(DA) |
-Macromolecule #3: RNA (5'-R(*(GTP)P*CP*CP*GP*A)-3')
| Macromolecule | Name: RNA (5'-R(*(GTP)P*CP*CP*GP*A)-3') / type: rna / ID: 3 |
|---|---|
| Source (natural) | Organism:  Escherichia phage T4 (virus) Escherichia phage T4 (virus) |
| Sequence | String: (GTP)CCGA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 Component:
Details: 20 mM HEPES pH 7.8, 100 mM NaCl, 10 mM MgCl2 and 2 mM DTT | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|
 Movie
Movie Controller
Controller























 Z
Z Y
Y X
X