+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of nucleosome-bound Sirtuin 6 deacetylase | |||||||||||||||
 Map data Map data | Main map from cisTEM, aligned to the common point of reference with the PDB and cryoSPARC map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Nucleosome / Sirt6 / aging / DNA damage / repair / deacetylation / diacylation / apo / chromatin / heterochromatin / GENE REGULATION / TRANSFERASE-DNA complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationhistone H3K56 deacetylase activity, NAD-dependent / histone H3K18 deacetylase activity, NAD-dependent / histone H3K9 deacetylase activity, NAD-dependent / protein delipidation / NAD+-protein-lysine ADP-ribosyltransferase activity / regulation of lipid catabolic process / ketone biosynthetic process / chromosome, subtelomeric region / NAD-dependent protein demyristoylase activity / NAD-dependent protein depalmitoylase activity ...histone H3K56 deacetylase activity, NAD-dependent / histone H3K18 deacetylase activity, NAD-dependent / histone H3K9 deacetylase activity, NAD-dependent / protein delipidation / NAD+-protein-lysine ADP-ribosyltransferase activity / regulation of lipid catabolic process / ketone biosynthetic process / chromosome, subtelomeric region / NAD-dependent protein demyristoylase activity / NAD-dependent protein depalmitoylase activity / NAD+-protein-arginine ADP-ribosyltransferase activity / positive regulation of protein localization to chromatin / DNA damage sensor activity / positive regulation of stem cell differentiation / positive regulation of blood vessel branching / NAD-dependent protein lysine deacetylase activity / transposable element silencing / positive regulation of chondrocyte proliferation / cardiac muscle cell differentiation / protein acetyllysine N-acetyltransferase / positive regulation of telomere maintenance / pericentric heterochromatin formation / histone deacetylase activity, NAD-dependent / protein deacetylation / negative regulation of D-glucose import / protein localization to site of double-strand break / TORC2 complex binding / negative regulation of glycolytic process / negative regulation of protein localization to chromatin / positive regulation of vascular endothelial cell proliferation / DNA repair-dependent chromatin remodeling / positive regulation of double-strand break repair / lncRNA binding / negative regulation of protein import into nucleus / regulation of double-strand break repair via homologous recombination / negative regulation of gene expression, epigenetic / positive regulation of stem cell population maintenance / regulation of protein secretion / positive regulation of stem cell proliferation / negative regulation of transcription elongation by RNA polymerase II / NAD+-protein ADP-ribosyltransferase activity / negative regulation of cellular senescence / site of DNA damage / regulation of lipid metabolic process / NAD+-protein poly-ADP-ribosyltransferase activity / Transferases; Glycosyltransferases; Pentosyltransferases / nucleosome binding / negative regulation of tumor necrosis factor-mediated signaling pathway / NAD+ binding / subtelomeric heterochromatin formation / positive regulation of fat cell differentiation / regulation of protein localization to plasma membrane / negative regulation of gluconeogenesis / protein localization to CENP-A containing chromatin / pericentric heterochromatin / Replacement of protamines by nucleosomes in the male pronucleus / CENP-A containing nucleosome / Packaging Of Telomere Ends / response to UV / Recognition and association of DNA glycosylase with site containing an affected purine / Cleavage of the damaged purine / Deposition of new CENPA-containing nucleosomes at the centromere / nucleosomal DNA binding / Transferases; Acyltransferases; Transferring groups other than aminoacyl groups / Recognition and association of DNA glycosylase with site containing an affected pyrimidine / Cleavage of the damaged pyrimidine / Inhibition of DNA recombination at telomere / Meiotic synapsis / nucleotidyltransferase activity / RNA Polymerase I Promoter Opening / Assembly of the ORC complex at the origin of replication / Regulation of endogenous retroelements by the Human Silencing Hub (HUSH) complex / DNA methylation / Condensation of Prophase Chromosomes / positive regulation of protein export from nucleus / SIRT1 negatively regulates rRNA expression / Chromatin modifications during the maternal to zygotic transition (MZT) / ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression / HCMV Late Events / innate immune response in mucosa / PRC2 methylates histones and DNA / determination of adult lifespan / Regulation of endogenous retroelements by KRAB-ZFP proteins / Defective pyroptosis / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / HDACs deacetylate histones / Nonhomologous End-Joining (NHEJ) / RNA Polymerase I Promoter Escape / lipopolysaccharide binding / Transcriptional regulation by small RNAs / protein destabilization / circadian regulation of gene expression / Formation of the beta-catenin:TCF transactivating complex / regulation of circadian rhythm / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / NoRC negatively regulates rRNA expression / base-excision repair / Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 / G2/M DNA damage checkpoint / positive regulation of insulin secretion Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) / | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.07 Å | |||||||||||||||
 Authors Authors | Chio US / Rechiche O / Bryll AR / Zhu J / Feldman JL / Peterson CL / Tan S / Armache J-P | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Cryo-EM structure of the human Sirtuin 6-nucleosome complex. Authors: Un Seng Chio / Othman Rechiche / Alysia R Bryll / Jiang Zhu / Erik M Leith / Jessica L Feldman / Craig L Peterson / Song Tan / Jean-Paul Armache /  Abstract: Sirtuin 6 (SIRT6) is a multifaceted protein deacetylase/deacylase and a major target for small-molecule modulators of longevity and cancer. In the context of chromatin, SIRT6 removes acetyl groups ...Sirtuin 6 (SIRT6) is a multifaceted protein deacetylase/deacylase and a major target for small-molecule modulators of longevity and cancer. In the context of chromatin, SIRT6 removes acetyl groups from histone H3 in nucleosomes, but the molecular basis for its nucleosomal substrate preference is unknown. Our cryo-electron microscopy structure of human SIRT6 in complex with the nucleosome shows that the catalytic domain of SIRT6 pries DNA from the nucleosomal entry-exit site and exposes the histone H3 N-terminal helix, while the SIRT6 zinc-binding domain binds to the histone acidic patch using an arginine anchor. In addition, SIRT6 forms an inhibitory interaction with the C-terminal tail of histone H2A. The structure provides insights into how SIRT6 can deacetylate both H3 K9 and H3 K56. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29735.map.gz emd_29735.map.gz | 91.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29735-v30.xml emd-29735-v30.xml emd-29735.xml emd-29735.xml | 36.6 KB 36.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29735.png emd_29735.png | 89.6 KB | ||
| Filedesc metadata |  emd-29735.cif.gz emd-29735.cif.gz | 8.7 KB | ||
| Others |  emd_29735_additional_1.map.gz emd_29735_additional_1.map.gz emd_29735_additional_2.map.gz emd_29735_additional_2.map.gz emd_29735_additional_3.map.gz emd_29735_additional_3.map.gz emd_29735_half_map_1.map.gz emd_29735_half_map_1.map.gz emd_29735_half_map_2.map.gz emd_29735_half_map_2.map.gz | 95.4 MB 91.5 MB 94.5 MB 10.4 MB 10.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29735 http://ftp.pdbj.org/pub/emdb/structures/EMD-29735 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29735 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29735 | HTTPS FTP |
-Validation report
| Summary document |  emd_29735_validation.pdf.gz emd_29735_validation.pdf.gz | 720.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29735_full_validation.pdf.gz emd_29735_full_validation.pdf.gz | 720.5 KB | Display | |
| Data in XML |  emd_29735_validation.xml.gz emd_29735_validation.xml.gz | 13.7 KB | Display | |
| Data in CIF |  emd_29735_validation.cif.gz emd_29735_validation.cif.gz | 16.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29735 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29735 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29735 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29735 | HTTPS FTP |
-Related structure data
| Related structure data |  8g57MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29735.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29735.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map from cisTEM, aligned to the common point of reference with the PDB and cryoSPARC map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
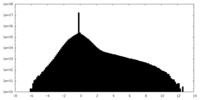
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Main map from cisTEM in original position
| File | emd_29735_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map from cisTEM in original position | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened main map from cisTEM, aligned to the...
| File | emd_29735_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened main map from cisTEM, aligned to the common point of reference with the PDB and cryoSPARC map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened and aligned main map from cryoSPARC
| File | emd_29735_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened and aligned main map from cryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1 from cisTEM
| File | emd_29735_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 from cisTEM | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 from cisTEM
| File | emd_29735_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 from cisTEM | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Sirt6 deacetylase bound to a nucleosome assembled with 172-bp 601...
| Entire | Name: Sirt6 deacetylase bound to a nucleosome assembled with 172-bp 601 Widom DNA |
|---|---|
| Components |
|
-Supramolecule #1: Sirt6 deacetylase bound to a nucleosome assembled with 172-bp 601...
| Supramolecule | Name: Sirt6 deacetylase bound to a nucleosome assembled with 172-bp 601 Widom DNA type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 300 kDa/nm |
-Macromolecule #1: NAD-dependent protein deacylase sirtuin-6
| Macromolecule | Name: NAD-dependent protein deacylase sirtuin-6 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Transferases; Acyltransferases; Transferring groups other than aminoacyl groups |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.708508 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSMSVNYAAG LSPYADKGKC GLPEIFDPPE ELERKVWELA RLVWQSSNVV FHTGAGISTA SGIPDFRGPH GVWTMEERGL APKFDTTFE SARPTQTHMA LVQLERVGLL RFLVSQNVDG LHVRSGFPRD KLAELHGNMF VEECAKCKTQ YVRDTVVGTM G LKATGRLC ...String: GSMSVNYAAG LSPYADKGKC GLPEIFDPPE ELERKVWELA RLVWQSSNVV FHTGAGISTA SGIPDFRGPH GVWTMEERGL APKFDTTFE SARPTQTHMA LVQLERVGLL RFLVSQNVDG LHVRSGFPRD KLAELHGNMF VEECAKCKTQ YVRDTVVGTM G LKATGRLC TVAKARGLRA CRGELRDTIL DWEDSLPDRD LALADEASRN ADLSITLGTS LQIRPSGNLP LATKRRGGRL VI VNLQPTK HDRHADLRIH GYVDEVMTRL MKHLGLEIPA WDGPRVLERA LPPLPRPPTP KLEPKEESPT RINGSIPAGP KQE PCAQHN GSEPASPKRE RPTSPAPHRP PKRVKAKAVP SKLN UniProtKB: NAD-dependent protein deacylase sirtuin-6 |
-Macromolecule #2: Histone H3
| Macromolecule | Name: Histone H3 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 15.004579 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TKQTARKSTG GKAPRKQLAT KAARKSAPAT GGVKKPHRYR PGTVALREIR RYQKSTELLI RKLPFQRLVR EIAQDFKTDL RFQSSAVMA LQEASEAYLV ALFEDTNLCA IHAKRVTIMP KDIQLARRIR GER UniProtKB: Histone H3 |
-Macromolecule #3: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 9.704396 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HRKVLRDNIQ GITKPAIRRL ARRGGVKRIS GLIYEETRGV LKVFLENVIR DAVTYTEHAK RKTVTAMDVV YALKRQGRTL YGFGG UniProtKB: Histone H4 |
-Macromolecule #4: Histone H2A type 1-B/E
| Macromolecule | Name: Histone H2A type 1-B/E / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.034355 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SGRGKQGGKA RAKAKTRSSR AGLQFPVGRV HRLLRKGNYS ERVGAGAPVY LAAVLEYLTA EILELAGNAA RDNKKTRIIP RHLQLAIRN DEELNKLLGR VTIAQGGVLP NIQAVLLPKK TESHHKAKGK UniProtKB: Histone H2A type 1-B/E |
-Macromolecule #5: Histone H2B type 1-J
| Macromolecule | Name: Histone H2B type 1-J / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.804045 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: PEPAKSAPAP KKGSKKAVTK AQKKDGKKRK RSRKESYSIY VYKVLKQVHP DTGISSKAMG IMNSFVNDIF ERIAGEASRL AHYNKRSTI TSREIQTAVR LLLPGELAKH AVSEGTKAVT KYTSAK UniProtKB: Histone H2B type 1-J |
-Macromolecule #6: DNA strand 1
| Macromolecule | Name: DNA strand 1 / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 46.541637 KDa |
| Sequence | String: (DT)(DG)(DC)(DA)(DC)(DA)(DG)(DG)(DA)(DT) (DG)(DT)(DA)(DT)(DA)(DT)(DA)(DT)(DC)(DT) (DG)(DA)(DC)(DA)(DC)(DG)(DT)(DG)(DC) (DC)(DT)(DG)(DG)(DA)(DG)(DA)(DC)(DT)(DA) (DG) (DG)(DG)(DA)(DG)(DT)(DA) ...String: (DT)(DG)(DC)(DA)(DC)(DA)(DG)(DG)(DA)(DT) (DG)(DT)(DA)(DT)(DA)(DT)(DA)(DT)(DC)(DT) (DG)(DA)(DC)(DA)(DC)(DG)(DT)(DG)(DC) (DC)(DT)(DG)(DG)(DA)(DG)(DA)(DC)(DT)(DA) (DG) (DG)(DG)(DA)(DG)(DT)(DA)(DA)(DT) (DC)(DC)(DC)(DC)(DT)(DT)(DG)(DG)(DC)(DG) (DG)(DT) (DT)(DA)(DA)(DA)(DA)(DC)(DG) (DC)(DG)(DG)(DG)(DG)(DG)(DA)(DC)(DA)(DG) (DC)(DG)(DC) (DG)(DT)(DA)(DC)(DG)(DT) (DG)(DC)(DG)(DT)(DT)(DT)(DA)(DA)(DG)(DC) (DG)(DG)(DT)(DG) (DC)(DT)(DA)(DG)(DA) (DG)(DC)(DT)(DG)(DT)(DC)(DT)(DA)(DC)(DG) (DA)(DC)(DC)(DA)(DA) (DT)(DT)(DG)(DA) (DG)(DC)(DG)(DG)(DC)(DC)(DT)(DC)(DG)(DG) (DC)(DA)(DC)(DC)(DG)(DG) (DG)(DA)(DT) (DT)(DC)(DT)(DC)(DG)(DA)(DT) |
-Macromolecule #7: DNA strand 2
| Macromolecule | Name: DNA strand 2 / type: dna / ID: 7 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 46.061348 KDa |
| Sequence | String: (DA)(DT)(DC)(DG)(DA)(DG)(DA)(DA)(DT)(DC) (DC)(DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA) (DG)(DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA) (DA)(DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA) (DG) (DA)(DC)(DA)(DG)(DC)(DT) ...String: (DA)(DT)(DC)(DG)(DA)(DG)(DA)(DA)(DT)(DC) (DC)(DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA) (DG)(DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA) (DA)(DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA) (DG) (DA)(DC)(DA)(DG)(DC)(DT)(DC)(DT) (DA)(DG)(DC)(DA)(DC)(DC)(DG)(DC)(DT)(DT) (DA)(DA) (DA)(DC)(DG)(DC)(DA)(DC)(DG) (DT)(DA)(DC)(DG)(DC)(DG)(DC)(DT)(DG)(DT) (DC)(DC)(DC) (DC)(DC)(DG)(DC)(DG)(DT) (DT)(DT)(DT)(DA)(DA)(DC)(DC)(DG)(DC)(DC) (DA)(DA)(DG)(DG) (DG)(DG)(DA)(DT)(DT) (DA)(DC)(DT)(DC)(DC)(DC)(DT)(DA)(DG)(DT) (DC)(DT)(DC)(DC)(DA) (DG)(DG)(DC)(DA) (DC)(DG)(DT)(DG)(DT)(DC)(DA)(DG)(DA)(DT) (DA)(DT)(DA)(DT)(DA)(DC) (DA)(DT)(DC) (DC)(DT)(DG)(DT)(DG)(DC)(DA) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 12.5 mM HEPES pH 7.5, 60 mM KCl, 1.5% glycerol, 1 mM DTT | |||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 1 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.2 kPa Details: Glass slides were wrapped with fresh parafilm. Tweezers were washed with ethanol, dried, and then used to pick grids from a grid box. Grids were carefully examined and placed on the parafilm- ...Details: Glass slides were wrapped with fresh parafilm. Tweezers were washed with ethanol, dried, and then used to pick grids from a grid box. Grids were carefully examined and placed on the parafilm-covered slides. These slides were then placed into the PelCO easyGLOW glow discharger, and treated there. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||
| Details | SIRT6 deacetylase bound to asymmetrical nucleosome with 172 DNA base-pairs |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 64.0 K / Max: 75.0 K |
| Specialist optics | Phase plate: OTHER / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 2 / Number real images: 11872 / Average exposure time: 3.3 sec. / Average electron dose: 50.0 e/Å2 Details: Data was collected in Super Resolution mode, thus the image size is 11520 (width) x 8184 (height) |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 2.2 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 81000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Details | UCSF Chimera was used for manual fitting of the models; It was then used for optimizing the fit by using option Fit in Map. Then Coot was used to analyze the fits, build and adjust the models into the existing densities, and refine parts of the model. Once the model has been built, validated and adjusted, we used phenix.real_space_refine to fix and improve the fit into the densities | ||||||||
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 79 | ||||||||
| Output model |  PDB-8g57: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)