+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Alzheimer's disease paired-helical filament in complex with PET tracer GTP-1 | |||||||||
 マップデータ マップデータ | ||||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | Neurodegeneration / Positron Emission Tomography / Filament / Alzheimer's disease / PROTEIN FIBRIL | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報plus-end-directed organelle transport along microtubule / histone-dependent DNA binding / negative regulation of establishment of protein localization to mitochondrion / positive regulation of protein localization to synapse / neurofibrillary tangle / microtubule lateral binding / axonal transport / main axon / phosphatidylinositol bisphosphate binding / tubulin complex ...plus-end-directed organelle transport along microtubule / histone-dependent DNA binding / negative regulation of establishment of protein localization to mitochondrion / positive regulation of protein localization to synapse / neurofibrillary tangle / microtubule lateral binding / axonal transport / main axon / phosphatidylinositol bisphosphate binding / tubulin complex / regulation of long-term synaptic depression / negative regulation of tubulin deacetylation / generation of neurons / regulation of chromosome organization / rRNA metabolic process / axonal transport of mitochondrion / regulation of mitochondrial fission / axon development / intracellular distribution of mitochondria / central nervous system neuron development / regulation of microtubule polymerization / microtubule polymerization / minor groove of adenine-thymine-rich DNA binding / lipoprotein particle binding / dynactin binding / negative regulation of mitochondrial membrane potential / glial cell projection / apolipoprotein binding / axolemma / protein polymerization / negative regulation of mitochondrial fission / Caspase-mediated cleavage of cytoskeletal proteins / regulation of microtubule polymerization or depolymerization / positive regulation of axon extension / neurofibrillary tangle assembly / regulation of microtubule cytoskeleton organization / regulation of cellular response to heat / Activation of AMPK downstream of NMDARs / synapse assembly / positive regulation of protein localization / supramolecular fiber organization / regulation of calcium-mediated signaling / cytoplasmic microtubule organization / somatodendritic compartment / cellular response to brain-derived neurotrophic factor stimulus / positive regulation of microtubule polymerization / axon cytoplasm / stress granule assembly / phosphatidylinositol binding / nuclear periphery / positive regulation of superoxide anion generation / protein phosphatase 2A binding / regulation of autophagy / cellular response to reactive oxygen species / astrocyte activation / Hsp90 protein binding / microglial cell activation / cellular response to nerve growth factor stimulus / synapse organization / response to lead ion / PKR-mediated signaling / protein homooligomerization / regulation of synaptic plasticity / SH3 domain binding / memory / microtubule cytoskeleton organization / cytoplasmic ribonucleoprotein granule / neuron projection development / microtubule cytoskeleton / cell-cell signaling / single-stranded DNA binding / protein-folding chaperone binding / actin binding / cell body / cellular response to heat / growth cone / double-stranded DNA binding / microtubule binding / protein-macromolecule adaptor activity / sequence-specific DNA binding / dendritic spine / amyloid fibril formation / microtubule / learning or memory / neuron projection / nuclear speck / membrane raft / axon / negative regulation of gene expression / neuronal cell body / dendrite / DNA damage response / protein kinase binding / enzyme binding / mitochondrion / DNA binding / RNA binding / extracellular region / identical protein binding / nucleus 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
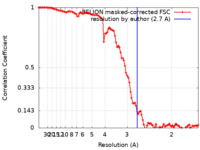
| 手法 | らせん対称体再構成法 / クライオ電子顕微鏡法 / 解像度: 2.7 Å | |||||||||
 データ登録者 データ登録者 | Merz GE / Tse E / Southworth DR | |||||||||
| 資金援助 |  米国, 2件 米国, 2件
| |||||||||
 引用 引用 |  ジャーナル: Nat Commun / 年: 2023 ジャーナル: Nat Commun / 年: 2023タイトル: Stacked binding of a PET ligand to Alzheimer's tau paired helical filaments. 著者: Gregory E Merz / Matthew J Chalkley / Sophia K Tan / Eric Tse / Joanne Lee / Stanley B Prusiner / Nick A Paras / William F DeGrado / Daniel R Southworth /  要旨: Accumulation of filamentous aggregates of tau protein in the brain is a pathological hallmark of Alzheimer's disease (AD) and many other neurodegenerative tauopathies. The filaments adopt disease- ...Accumulation of filamentous aggregates of tau protein in the brain is a pathological hallmark of Alzheimer's disease (AD) and many other neurodegenerative tauopathies. The filaments adopt disease-specific cross-β amyloid conformations that self-propagate and are implicated in neuronal loss. Development of molecular diagnostics and therapeutics is of critical importance. However, mechanisms of small molecule binding to the amyloid core is poorly understood. We used cryo-electron microscopy to determine a 2.7 Å structure of AD patient-derived tau paired-helical filaments bound to the PET ligand GTP-1. The compound is bound stoichiometrically at a single site along an exposed cleft of each protofilament in a stacked arrangement matching the fibril symmetry. Multiscale modeling reveals pi-pi aromatic interactions that pair favorably with the small molecule-protein contacts, supporting high specificity and affinity for the AD tau conformation. This binding mode offers critical insight into designing compounds to target different amyloid folds found across neurodegenerative diseases. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_29458.map.gz emd_29458.map.gz | 7.2 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-29458-v30.xml emd-29458-v30.xml emd-29458.xml emd-29458.xml | 16.3 KB 16.3 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_29458_fsc.xml emd_29458_fsc.xml | 10.2 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_29458.png emd_29458.png | 70.2 KB | ||
| Filedesc metadata |  emd-29458.cif.gz emd-29458.cif.gz | 5.6 KB | ||
| その他 |  emd_29458_half_map_1.map.gz emd_29458_half_map_1.map.gz emd_29458_half_map_2.map.gz emd_29458_half_map_2.map.gz | 71.3 MB 71.3 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29458 http://ftp.pdbj.org/pub/emdb/structures/EMD-29458 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29458 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29458 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_29458_validation.pdf.gz emd_29458_validation.pdf.gz | 775.7 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_29458_full_validation.pdf.gz emd_29458_full_validation.pdf.gz | 775.2 KB | 表示 | |
| XML形式データ |  emd_29458_validation.xml.gz emd_29458_validation.xml.gz | 17.5 KB | 表示 | |
| CIF形式データ |  emd_29458_validation.cif.gz emd_29458_validation.cif.gz | 22.8 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29458 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29458 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29458 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29458 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8fugMC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_29458.map.gz / 形式: CCP4 / 大きさ: 91.1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_29458.map.gz / 形式: CCP4 / 大きさ: 91.1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.834 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-ハーフマップ: #1
| ファイル | emd_29458_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: #2
| ファイル | emd_29458_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Alzheimer's tau paired helical filament in complex with PET ligan...
| 全体 | 名称: Alzheimer's tau paired helical filament in complex with PET ligand GTP-1 |
|---|---|
| 要素 |
|
-超分子 #1: Alzheimer's tau paired helical filament in complex with PET ligan...
| 超分子 | 名称: Alzheimer's tau paired helical filament in complex with PET ligand GTP-1 タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1 |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: Microtubule-associated protein tau
| 分子 | 名称: Microtubule-associated protein tau / タイプ: protein_or_peptide / ID: 1 / コピー数: 23 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 7.940141 KDa |
| 配列 | 文字列: VQIVYKPVDL SKVTSKCGSL GNIHHKPGGG QVEVKSEKLD FKDRVQSKIG SLDNITHVPG GGNKKIETHK LTF UniProtKB: Microtubule-associated protein tau |
-分子 #2: (5S)-2-[4-(2-fluoroethyl)piperidin-1-yl]pyrimido[1,2-a]benzimidazole
| 分子 | 名称: (5S)-2-[4-(2-fluoroethyl)piperidin-1-yl]pyrimido[1,2-a]benzimidazole タイプ: ligand / ID: 2 / コピー数: 23 / 式: Y9H |
|---|---|
| 分子量 | 理論値: 298.358 Da |
| Chemical component information | 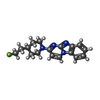 ChemComp-Y9H: |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | らせん対称体再構成法 |
| 試料の集合状態 | filament |
- 試料調製
試料調製
| 緩衝液 | pH: 7.4 構成要素:
詳細: 20 mM Tris-HCl, pH 7.4, 100 mM NaCl | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| グリッド | モデル: Quantifoil R1.2/1.3 / 材質: GOLD / メッシュ: 200 / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: HOLEY / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 時間: 120 sec. | |||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 277 K / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 特殊光学系 | エネルギーフィルター - 名称: GIF Bioquantum / エネルギーフィルター - スリット幅: 20 eV |
| 撮影 | フィルム・検出器のモデル: GATAN K3 (6k x 4k) / 実像数: 15160 / 平均電子線量: 46.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): 1.8 µm / 最小 デフォーカス(公称値): 0.8 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)