[English] 日本語
 Yorodumi
Yorodumi- EMDB-29311: Cryo-EM structure of RNase-untreated RESC-C in trypanosomal RNA e... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of RNase-untreated RESC-C in trypanosomal RNA editing | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | HEAT repeat / trypanosoma / RNA editing substrate binding complex / gRNA / RNA BINDING PROTEIN-RNA complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of mitochondrial mRNA stability / mitochondrial mRNA processing / RNA modification / mitochondrial mRNA editing complex / ribonucleoprotein granule / RNA metabolic process / mitochondrial RNA modification / mitochondrial RNA processing / cytidine to uridine editing / kinetoplast ...regulation of mitochondrial mRNA stability / mitochondrial mRNA processing / RNA modification / mitochondrial mRNA editing complex / ribonucleoprotein granule / RNA metabolic process / mitochondrial RNA modification / mitochondrial RNA processing / cytidine to uridine editing / kinetoplast / 2-oxoglutarate-dependent dioxygenase activity / mRNA stabilization / post-transcriptional regulation of gene expression / RNA processing / mitochondrial matrix / mRNA binding / mitochondrion / RNA binding / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
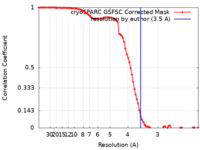
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | ||||||||||||
 Authors Authors | Liu S / Wang H / Li X / Zhang F / Lee JKJ / Li Z / Yu C / Zhao X / Hu JJ / Suematsu T ...Liu S / Wang H / Li X / Zhang F / Lee JKJ / Li Z / Yu C / Zhao X / Hu JJ / Suematsu T / Alvarez-Cabrera AL / Liu Q / Zhang L / Huang L / Aphasizheva I / Aphasizhev R / Zhou ZH | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Science / Year: 2023 Journal: Science / Year: 2023Title: Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing. Authors: Shiheng Liu / Hong Wang / Xiaorun Li / Fan Zhang / Jane K J Lee / Zihang Li / Clinton Yu / Jason J Hu / Xiaojing Zhao / Takuma Suematsu / Ana L Alvarez-Cabrera / Qiushi Liu / Liye Zhang / ...Authors: Shiheng Liu / Hong Wang / Xiaorun Li / Fan Zhang / Jane K J Lee / Zihang Li / Clinton Yu / Jason J Hu / Xiaojing Zhao / Takuma Suematsu / Ana L Alvarez-Cabrera / Qiushi Liu / Liye Zhang / Lan Huang / Inna Aphasizheva / Ruslan Aphasizhev / Z Hong Zhou /   Abstract: In , the editosome, composed of RNA-editing substrate-binding complex (RESC) and RNA-editing catalytic complex (RECC), orchestrates guide RNA (gRNA)-programmed editing to recode cryptic mitochondrial ...In , the editosome, composed of RNA-editing substrate-binding complex (RESC) and RNA-editing catalytic complex (RECC), orchestrates guide RNA (gRNA)-programmed editing to recode cryptic mitochondrial transcripts into messenger RNAs (mRNAs). The mechanism of information transfer from gRNA to mRNA is unclear owing to a lack of high-resolution structures for these complexes. With cryo-electron microscopy and functional studies, we have captured gRNA-stabilizing RESC-A and gRNA-mRNA-binding RESC-B and RESC-C particles. RESC-A sequesters gRNA termini, thus promoting hairpin formation and blocking mRNA access. The conversion of RESC-A into RESC-B or -C unfolds gRNA and allows mRNA selection. The ensuing gRNA-mRNA duplex protrudes from RESC-B, likely exposing editing sites to RECC-catalyzed cleavage, uridine insertion or deletion, and ligation. Our work reveals a remodeling event facilitating gRNA-mRNA hybridization and assembly of a macromolecular substrate for the editosome's catalytic modality. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29311.map.gz emd_29311.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29311-v30.xml emd-29311-v30.xml emd-29311.xml emd-29311.xml | 23.6 KB 23.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29311_fsc.xml emd_29311_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_29311.png emd_29311.png | 118.1 KB | ||
| Filedesc metadata |  emd-29311.cif.gz emd-29311.cif.gz | 7.5 KB | ||
| Others |  emd_29311_half_map_1.map.gz emd_29311_half_map_1.map.gz emd_29311_half_map_2.map.gz emd_29311_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29311 http://ftp.pdbj.org/pub/emdb/structures/EMD-29311 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29311 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29311 | HTTPS FTP |
-Validation report
| Summary document |  emd_29311_validation.pdf.gz emd_29311_validation.pdf.gz | 832.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29311_full_validation.pdf.gz emd_29311_full_validation.pdf.gz | 832.2 KB | Display | |
| Data in XML |  emd_29311_validation.xml.gz emd_29311_validation.xml.gz | 16.5 KB | Display | |
| Data in CIF |  emd_29311_validation.cif.gz emd_29311_validation.cif.gz | 21.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29311 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29311 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29311 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29311 | HTTPS FTP |
-Related structure data
| Related structure data |  8fnfMC  8fn4C  8fn6C  8fncC  8fniC  8fnkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29311.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29311.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29311_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29311_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : RESC5-tagged isolate without RNase treatment
| Entire | Name: RESC5-tagged isolate without RNase treatment |
|---|---|
| Components |
|
-Supramolecule #1: RESC5-tagged isolate without RNase treatment
| Supramolecule | Name: RESC5-tagged isolate without RNase treatment / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: CTS-tagged RESC5 purified from RNase-untreated mitochondrial extract by tandem affinity procedure |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: mRNA
| Macromolecule | Name: mRNA / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 6.45596 KDa |
| Sequence | String: UAAUAGAAUA AGAUAUAAAA |
-Macromolecule #2: gRNA
| Macromolecule | Name: gRNA / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 4.968896 KDa |
| Sequence | String: UUUUUUUAAA UAAUUU |
-Macromolecule #3: Mitochondrial RNA binding protein
| Macromolecule | Name: Mitochondrial RNA binding protein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.727539 KDa |
| Sequence | String: MLRHTSRNNA LHAFVRSPHY RTIPSAGPNG IVVNRDMLVH QFRDFYKTLQ HCSLVDKVHL MSERPSVEAL RVADQMVSIG ATFLEMPLT GMEHRATEFM ESMRYVRGAG GPSTLASYLQ DTENCRCNSG DVVCLPNGIA VGHGPRTNAV AHTTLKQLFE V KDDQFSFD ...String: MLRHTSRNNA LHAFVRSPHY RTIPSAGPNG IVVNRDMLVH QFRDFYKTLQ HCSLVDKVHL MSERPSVEAL RVADQMVSIG ATFLEMPLT GMEHRATEFM ESMRYVRGAG GPSTLASYLQ DTENCRCNSG DVVCLPNGIA VGHGPRTNAV AHTTLKQLFE V KDDQFSFD VFTLEQEGDA PPLGDYFGFA GSNVLLTWKD EHGLLAVDQY QQKQPHTEMN VVYLEPGCHF LSFYGVDHTI DV LVQKGYE RSMDSIAAAG LNPIPVQWSE MDKLGISMRA AVLPLKFFKA NVGGMLSRNK SRGARWQTHQ LQK UniProtKB: Mitochondrial RNA binding protein |
-Macromolecule #4: RAP domain-containing protein
| Macromolecule | Name: RAP domain-containing protein / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 57.823484 KDa |
| Sequence | String: MRSALRRCIL RHQGCLRMKQ SLSAFPTVVT GMTRHQGNSL IGTTHGAELS LAGDPQSVSH LSARNIATEA LQMKKLHQER GGNPMLAQQ ARRVLFATSI AGQNLDARSV ALLLNTAVYF GMESDAKLVR ECIDYCLKND KLITVDVLPI VVTACATLKS R DAREVIEM ...String: MRSALRRCIL RHQGCLRMKQ SLSAFPTVVT GMTRHQGNSL IGTTHGAELS LAGDPQSVSH LSARNIATEA LQMKKLHQER GGNPMLAQQ ARRVLFATSI AGQNLDARSV ALLLNTAVYF GMESDAKLVR ECIDYCLKND KLITVDVLPI VVTACATLKS R DAREVIEM QAQKAARNAK FLDAKDVTNI ISAFSKTGIN HEKLFAFLSR RVQTLARVGE FEAAHLVILA NAFSRLRYRD KF LFGAIAR RAMSLRERVT VNELVPLIVA FSKIGLKDPK LSKRFATKAM EYVDQMNAEQ VASMFMAFAY FGIRYDQLFG VLT NRAVEL IDEFNAQYIS TTLNAFQRIG INNPELFDNL AERALAVVQD HDARDISKTV TALAHFGLKD EELFKRLASH AASI ADQFD AMGLVNTAHA FARTNFLQQD MAVALSERSV YVCRLLDAGE TRRLLWALAK FQVRDPKILT PVFNRCLALH YDFFA DPTG SEEIEEIFDF YGPNFCPPLY QLYISRGSTP QA UniProtKB: Guide RNA binding protein |
-Macromolecule #5: RxLR effector protein
| Macromolecule | Name: RxLR effector protein / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.669676 KDa |
| Sequence | String: MRSSRGILFL SGAFAIRGMS AYHSYQRLDT VSHTSKVYSL QMQRQTVHFT PITRLGVEAT ANPTTATNAT GQTGDGDGAT ALDVAMRVN KLKRLHQTGG GPSGKKQVEL DAWRDLNNLT EAQINSAEGK AVSLLLNSWA YFAKYWEKGA EGPSASLSEV T PSNDSSSA GEHGTQ UniProtKB: Mitochondrial RNA binding complex 1 subunit |
-Macromolecule #6: Mitochondrial RNA binding complex 1 subunit
| Macromolecule | Name: Mitochondrial RNA binding complex 1 subunit / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 61.109809 KDa |
| Sequence | String: MLNVLSSTAS AALATVVVAR PSALHLIFER CKLNLVEFTA QDVYQICTTA YNMDTLGMLQ DPDFMRGLHD AFRRSDQTVI SPFQANLIA DTFRKVGINS MPKEVSVPEE DAISPESLIL VLRNMNITKQ RDERKINEVL KLMFPILDEF SPTQLSLTVT E LARLKSTN ...String: MLNVLSSTAS AALATVVVAR PSALHLIFER CKLNLVEFTA QDVYQICTTA YNMDTLGMLQ DPDFMRGLHD AFRRSDQTVI SPFQANLIA DTFRKVGINS MPKEVSVPEE DAISPESLIL VLRNMNITKQ RDERKINEVL KLMFPILDEF SPTQLSLTVT E LARLKSTN ADFVGKLAKR IMEYNDDLSA LDISSAAVSL AYCPGISHNI LYRMMQIVEE RMGEFQPEDY INVLHALNTL GP KFVNTFR KIVECGLQHV ENMDAVTLTN YMVCFSTMDY KQREHIDIYA DALVEVATDL SEKDLVMAFI ALQRLRLLSD TMF GTMASC VIRYAAKMDP RNIAPIMDIC STVPHASDHL MKVLMDRAVE CTRILTANQL GDILDILGLY PPAREHPLVQ LFGK QARLR LDLMGPDALA NATRGLANLG YADPEYYAQA AETGFRYGFK DWTLLEPMLM GLSITGQCPP TMVRVLGSHI APMAR SMSL MEIERANRYL RRLGCEDDFV YKAMASRVLQ FVKEVTPEMP EDLQVLLQRG AVEPGAAPGV M UniProtKB: Mitochondrial RNA binding complex 1 subunit |
-Macromolecule #7: RAP domain-containing protein
| Macromolecule | Name: RAP domain-containing protein / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 60.563676 KDa |
| Sequence | String: MRRRVVLCCQ DVGSLLSSKH SVHSGIGYHE RVFSRNLLYR RYPVVTVLPK AGFTVLDTKR WIASSGPPVT GSPLSPVTNP SLNVGTGGG EAVAMEGPLP VSYSPGSGVN GSLPVTSTAI TAHCDVLSEC VAKADELAVQ LKAQNALSAS AEILTQEGME E FVEELKTS ...String: MRRRVVLCCQ DVGSLLSSKH SVHSGIGYHE RVFSRNLLYR RYPVVTVLPK AGFTVLDTKR WIASSGPPVT GSPLSPVTNP SLNVGTGGG EAVAMEGPLP VSYSPGSGVN GSLPVTSTAI TAHCDVLSEC VAKADELAVQ LKAQNALSAS AEILTQEGME E FVEELKTS ATNEMTALVK QMQTTPLLQR AGMHELRRTL YYTTSLKERD WLEEKQYTAA MRMLTVEVLR RDGDGVLSAD DV LYVTTHV VTANFYNRHL WNRMEKSLLK FSNYENIDMS SVKAFSTRLF KTRRGCAKET LDIRRKVLLA MSRRVGVLAN DFD LPSLLG VLQCYTVHDL TPFHLEPLAI RATNHVGDFT PHECATLAHV LRKWRTMRLE VCERLVERIC TSDQLTHHMA NAAM IAIRT CFNQVSDGGR NAMNAEPTRQ KLRAMGEQIG CRLDEVEYPA LPVILSILDV VVTLKIYVPK KCLQVIFSQA NDMVA IVME QKDDLVDPKT GKRVRPITAE EGRQLQALLS HYGNDLAPEL SQRMKEAFRE GVLPDEASL UniProtKB: Mitochondrial RNA binding complex 1 subunit |
-Macromolecule #8: Phytanoyl-CoA dioxygenase family protein
| Macromolecule | Name: Phytanoyl-CoA dioxygenase family protein / type: protein_or_peptide / ID: 8 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.293148 KDa |
| Sequence | String: MRSGRKLGCF TNRLRLPFFS PCSQITALTA SHRCKSYVLK FLRGQLPEDL KDVNGALGCL YGTLPDVDEF GQFVISPDVV NSFHQFGYV KMPIPVLDHQ QIDKLADEVN ELANNVEHHP KTERLYATSL ADLTGGPLFF CQGQWRAAWG MHDLIYLPTI T VAASQILN ...String: MRSGRKLGCF TNRLRLPFFS PCSQITALTA SHRCKSYVLK FLRGQLPEDL KDVNGALGCL YGTLPDVDEF GQFVISPDVV NSFHQFGYV KMPIPVLDHQ QIDKLADEVN ELANNVEHHP KTERLYATSL ADLTGGPLFF CQGQWRAAWG MHDLIYLPTI T VAASQILN NSLVRLWYDE VFMKAARTGP CVPWQQNYAR WQHTKPVNHV TVMIALDTMN KDRGAPCLVP GSHRWREGGL LP PVSYDPT KDEAHQLNTI WEIINEEEGE MLMDTPPVTV DLRRGEALLI HPLTLFATHG NRSLDAVRCC FIHYMGEKTY AVQ NGPLLP HTTKFQADAM IQGPFYPVVF DPAMTEELTM LPTAPSEEEA UniProtKB: Phytanoyl-CoA dioxygenase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)