[English] 日本語
 Yorodumi
Yorodumi- EMDB-29316: Cryo-EM structure of RNase-untreated RESC-B in trypanosomal RNA e... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of RNase-untreated RESC-B in trypanosomal RNA editing | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | HEAT repeat / trypanosoma / RNA editing substrate binding complex / gRNA / RNA BINDING PROTEIN-RNA complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of mitochondrial mRNA stability / mitochondrial mRNA processing / RNA modification / mitochondrial mRNA editing complex / RNA metabolic process / ribonucleoprotein granule / mitochondrial RNA modification / mitochondrial RNA processing / cytidine to uridine editing / kinetoplast ...regulation of mitochondrial mRNA stability / mitochondrial mRNA processing / RNA modification / mitochondrial mRNA editing complex / RNA metabolic process / ribonucleoprotein granule / mitochondrial RNA modification / mitochondrial RNA processing / cytidine to uridine editing / kinetoplast / cytoplasmic side of mitochondrial outer membrane / mRNA stabilization / post-transcriptional regulation of gene expression / RNA processing / mitochondrial matrix / ribonucleoprotein complex / mRNA binding / mitochondrion / RNA binding / nucleus / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | ||||||||||||
 Authors Authors | Liu S / Wang H / Li X / Zhang F / Lee JKJ / Li Z / Yu C / Zhao X / Hu JJ / Suematsu T ...Liu S / Wang H / Li X / Zhang F / Lee JKJ / Li Z / Yu C / Zhao X / Hu JJ / Suematsu T / Alvarez-Cabrera AL / Liu Q / Zhang L / Huang L / Aphasizheva I / Aphasizhev R / Zhou ZH | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Science / Year: 2023 Journal: Science / Year: 2023Title: Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing. Authors: Shiheng Liu / Hong Wang / Xiaorun Li / Fan Zhang / Jane K J Lee / Zihang Li / Clinton Yu / Jason J Hu / Xiaojing Zhao / Takuma Suematsu / Ana L Alvarez-Cabrera / Qiushi Liu / Liye Zhang / ...Authors: Shiheng Liu / Hong Wang / Xiaorun Li / Fan Zhang / Jane K J Lee / Zihang Li / Clinton Yu / Jason J Hu / Xiaojing Zhao / Takuma Suematsu / Ana L Alvarez-Cabrera / Qiushi Liu / Liye Zhang / Lan Huang / Inna Aphasizheva / Ruslan Aphasizhev / Z Hong Zhou /   Abstract: In , the editosome, composed of RNA-editing substrate-binding complex (RESC) and RNA-editing catalytic complex (RECC), orchestrates guide RNA (gRNA)-programmed editing to recode cryptic mitochondrial ...In , the editosome, composed of RNA-editing substrate-binding complex (RESC) and RNA-editing catalytic complex (RECC), orchestrates guide RNA (gRNA)-programmed editing to recode cryptic mitochondrial transcripts into messenger RNAs (mRNAs). The mechanism of information transfer from gRNA to mRNA is unclear owing to a lack of high-resolution structures for these complexes. With cryo-electron microscopy and functional studies, we have captured gRNA-stabilizing RESC-A and gRNA-mRNA-binding RESC-B and RESC-C particles. RESC-A sequesters gRNA termini, thus promoting hairpin formation and blocking mRNA access. The conversion of RESC-A into RESC-B or -C unfolds gRNA and allows mRNA selection. The ensuing gRNA-mRNA duplex protrudes from RESC-B, likely exposing editing sites to RECC-catalyzed cleavage, uridine insertion or deletion, and ligation. Our work reveals a remodeling event facilitating gRNA-mRNA hybridization and assembly of a macromolecular substrate for the editosome's catalytic modality. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29316.map.gz emd_29316.map.gz | 398.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29316-v30.xml emd-29316-v30.xml emd-29316.xml emd-29316.xml | 28.7 KB 28.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29316_fsc.xml emd_29316_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_29316.png emd_29316.png | 92.7 KB | ||
| Filedesc metadata |  emd-29316.cif.gz emd-29316.cif.gz | 9.1 KB | ||
| Others |  emd_29316_half_map_1.map.gz emd_29316_half_map_1.map.gz emd_29316_half_map_2.map.gz emd_29316_half_map_2.map.gz | 391.1 MB 391.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29316 http://ftp.pdbj.org/pub/emdb/structures/EMD-29316 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29316 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29316 | HTTPS FTP |
-Related structure data
| Related structure data |  8fnkMC  8fn4C  8fn6C  8fncC  8fnfC  8fniC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29316.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29316.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29316_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
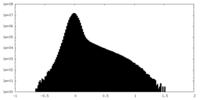
| Projections & Slices |
| ||||||||||||
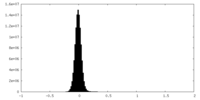
| Density Histograms |
-Half map: #1
| File | emd_29316_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
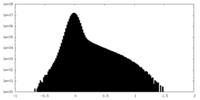
| Projections & Slices |
| ||||||||||||
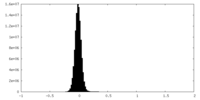
| Density Histograms |
- Sample components
Sample components
+Entire : RESC5-tagged isolate without RNase treatment
+Supramolecule #1: RESC5-tagged isolate without RNase treatment
+Macromolecule #1: mRNA
+Macromolecule #2: gRNA
+Macromolecule #3: RNA-editing substrate-binding complex protein 5 (RESC5)
+Macromolecule #4: RNA-editing substrate-binding complex protein 6 (RESC6)
+Macromolecule #5: RNA-editing substrate-binding complex protein 7 (RESC7)
+Macromolecule #6: RNA-editing substrate-binding complex protein 8 (RESC8)
+Macromolecule #7: RNA-editing substrate-binding complex protein 9 (RESC9)
+Macromolecule #8: RNA-editing substrate-binding complex protein 10 (RESC10)
+Macromolecule #9: RNA-editing substrate-binding complex protein 11 (RESC11)
+Macromolecule #10: RNA-editing substrate-binding complex protein 13 (RESC13)
+Macromolecule #11: RNA-editing substrate-binding complex protein 14 (RESC14)
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)