+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
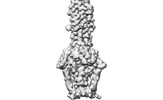
| Title | Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | VIRAL PROTEIN | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | ||||||||||||
 Authors Authors | Zhang J / Shi W / Cai YF / Zhu HS / Peng HQ / Voyer J / Volloch SR / Cao H / Mayer ML / Song KK ...Zhang J / Shi W / Cai YF / Zhu HS / Peng HQ / Voyer J / Volloch SR / Cao H / Mayer ML / Song KK / Xu C / Lu JM / Chen B | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane. Authors: Wei Shi / Yongfei Cai / Haisun Zhu / Hanqin Peng / Jewel Voyer / Sophia Rits-Volloch / Hong Cao / Megan L Mayer / Kangkang Song / Chen Xu / Jianming Lu / Jun Zhang / Bing Chen /  Abstract: The entry of SARS-CoV-2 into host cells depends on the refolding of the virus-encoded spike protein from a prefusion conformation, which is metastable after cleavage, to a lower-energy stable ...The entry of SARS-CoV-2 into host cells depends on the refolding of the virus-encoded spike protein from a prefusion conformation, which is metastable after cleavage, to a lower-energy stable postfusion conformation. This transition overcomes kinetic barriers for fusion of viral and target cell membranes. Here we report a cryogenic electron microscopy (cryo-EM) structure of the intact postfusion spike in a lipid bilayer that represents the single-membrane product of the fusion reaction. The structure provides structural definition of the functionally critical membrane-interacting segments, including the fusion peptide and transmembrane anchor. The internal fusion peptide forms a hairpin-like wedge that spans almost the entire lipid bilayer and the transmembrane segment wraps around the fusion peptide at the last stage of membrane fusion. These results advance our understanding of the spike protein in a membrane environment and may guide development of intervention strategies. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29017.map.gz emd_29017.map.gz | 698.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29017-v30.xml emd-29017-v30.xml emd-29017.xml emd-29017.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29017.png emd_29017.png | 36.7 KB | ||
| Others |  emd_29017_half_map_1.map.gz emd_29017_half_map_1.map.gz emd_29017_half_map_2.map.gz emd_29017_half_map_2.map.gz | 763.8 MB 763.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29017 http://ftp.pdbj.org/pub/emdb/structures/EMD-29017 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29017 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29017 | HTTPS FTP |
-Validation report
| Summary document |  emd_29017_validation.pdf.gz emd_29017_validation.pdf.gz | 729.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29017_full_validation.pdf.gz emd_29017_full_validation.pdf.gz | 729.3 KB | Display | |
| Data in XML |  emd_29017_validation.xml.gz emd_29017_validation.xml.gz | 20.2 KB | Display | |
| Data in CIF |  emd_29017_validation.cif.gz emd_29017_validation.cif.gz | 24.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29017 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29017 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29017 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29017 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29017.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29017.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_29017_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
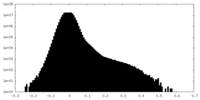
| Density Histograms |
-Half map: #2
| File | emd_29017_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
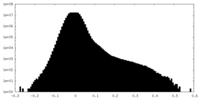
| Density Histograms |
- Sample components
Sample components
-Entire : local map of SRAS-CoV-2 postfusion spike protein in nanodisc from...
| Entire | Name: local map of SRAS-CoV-2 postfusion spike protein in nanodisc from data set I |
|---|---|
| Components |
|
-Supramolecule #1: local map of SRAS-CoV-2 postfusion spike protein in nanodisc from...
| Supramolecule | Name: local map of SRAS-CoV-2 postfusion spike protein in nanodisc from data set I type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: local map of SARS-CoV-2 postfusion spike protein in nanodisc from data set I |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 210 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.01 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 14-1211 / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Protocol: AB INITIO MODEL |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)