+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of TnsC oligomer in type I-B CAST system | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TnsC / CAST / DNA BINDING PROTEIN | |||||||||
| Function / homology | : / AAA domain / ATP hydrolysis activity / P-loop containing nucleoside triphosphate hydrolase / ORC1/DEAH AAA+ ATPase domain-containing protein Function and homology information Function and homology information | |||||||||
| Biological species |  Peltigera membranacea (fungus) / Peltigera membranacea (fungus) /  Nostoc sp. 'Peltigera membranacea cyanobiont' 210A (bacteria) Nostoc sp. 'Peltigera membranacea cyanobiont' 210A (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Chang L / Wang S | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Molecular mechanism for Tn7-like transposon recruitment by a type I-B CRISPR effector. Authors: Shukun Wang / Clinton Gabel / Romana Siddique / Thomas Klose / Leifu Chang /  Abstract: Tn7-like transposons have co-opted CRISPR-Cas systems to facilitate the movement of their own DNA. These CRISPR-associated transposons (CASTs) are promising tools for programmable gene knockin. A key ...Tn7-like transposons have co-opted CRISPR-Cas systems to facilitate the movement of their own DNA. These CRISPR-associated transposons (CASTs) are promising tools for programmable gene knockin. A key feature of CASTs is their ability to recruit Tn7-like transposons to nuclease-deficient CRISPR effectors. However, how Tn7-like transposons are recruited by diverse CRISPR effectors remains poorly understood. Here, we present the cryo-EM structure of a recruitment complex comprising the Cascade complex, TniQ, TnsC, and the target DNA in the type I-B CAST from Peltigera membranacea cyanobiont 210A. Target DNA recognition by Cascade induces conformational changes in Cas6 and primes TniQ recruitment through its C-terminal domain. The N-terminal domain of TniQ is bound to the seam region of the TnsC spiral heptamer. Our findings provide insights into the diverse mechanisms for the recruitment of Tn7-like transposons to CRISPR effectors and will aid in the development of CASTs as gene knockin tools. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28998.map.gz emd_28998.map.gz | 28.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28998-v30.xml emd-28998-v30.xml emd-28998.xml emd-28998.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28998.png emd_28998.png | 103.4 KB | ||
| Filedesc metadata |  emd-28998.cif.gz emd-28998.cif.gz | 5.2 KB | ||
| Others |  emd_28998_half_map_1.map.gz emd_28998_half_map_1.map.gz emd_28998_half_map_2.map.gz emd_28998_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28998 http://ftp.pdbj.org/pub/emdb/structures/EMD-28998 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28998 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28998 | HTTPS FTP |
-Validation report
| Summary document |  emd_28998_validation.pdf.gz emd_28998_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28998_full_validation.pdf.gz emd_28998_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_28998_validation.xml.gz emd_28998_validation.xml.gz | 12.2 KB | Display | |
| Data in CIF |  emd_28998_validation.cif.gz emd_28998_validation.cif.gz | 14.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28998 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28998 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28998 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28998 | HTTPS FTP |
-Related structure data
| Related structure data |  8fcxMC  8fcjC  8fcuC  8fcvC  8fcwC  8fd2C  8fd3C  8ff4C  8ff5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28998.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28998.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.054 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_28998_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
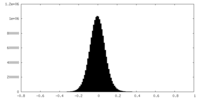
| Density Histograms |
-Half map: #1
| File | emd_28998_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
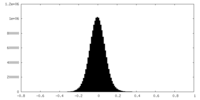
| Density Histograms |
- Sample components
Sample components
-Entire : TnsC heptamer
| Entire | Name: TnsC heptamer |
|---|---|
| Components |
|
-Supramolecule #1: TnsC heptamer
| Supramolecule | Name: TnsC heptamer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Peltigera membranacea (fungus) Peltigera membranacea (fungus) |
-Macromolecule #1: TnsC
| Macromolecule | Name: TnsC / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nostoc sp. 'Peltigera membranacea cyanobiont' 210A (bacteria) Nostoc sp. 'Peltigera membranacea cyanobiont' 210A (bacteria) |
| Molecular weight | Theoretical: 43.521934 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTKSTGFPLE LLTRPATERL AYFENYTVAH PRLKEVYEIL MRTIAEPAGA SFIFVYGASG VGKTTLRLRV EQKLTELALP KLESDRARV PVVGIEAIAP ESRYFNWKEY YTRALITLEE PLIDHKFDYG VRGISRDNFG KINVESKVVA PALRRALENA L IHRHPDVF ...String: MTKSTGFPLE LLTRPATERL AYFENYTVAH PRLKEVYEIL MRTIAEPAGA SFIFVYGASG VGKTTLRLRV EQKLTELALP KLESDRARV PVVGIEAIAP ESRYFNWKEY YTRALITLEE PLIDHKFDYG VRGISRDNFG KINVESKVVA PALRRALENA L IHRHPDVF FVDEAQHFGK VASGYKLQDQ LDCLKSLANM TGILHCLLGT YELLTFRNLS GQLSRRSVDI HFRRYCADSP ED VQAFKSV LLTFQQHLPL AETPNLVDHW EYFYERTLGC IGTLKDWLKR VLSDALDREA TTITLKDLQK RALSVAQCQK MFK EIQEGE RQLSETEADV QNLRSALGLG AKPIVLPEET PKTTRPPGKV GKRKPKRDPI GVQQDVS UniProtKB: ORC1/DEAH AAA+ ATPase domain-containing protein |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 5 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 5 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.04 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 193399 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller



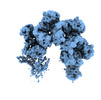








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)