[English] 日本語
 Yorodumi
Yorodumi- EMDB-28980: Cryo-EM structure of Cascade-DNA (P23) complex in type I-B CAST system -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Cascade-DNA (P23) complex in type I-B CAST system | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR / DNA recognition / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Peltigera membranacea (fungus) / Peltigera membranacea (fungus) /  Nostoc sp. 'Peltigera membranacea cyanobiont' 210A (bacteria) / synthetic construct (others) Nostoc sp. 'Peltigera membranacea cyanobiont' 210A (bacteria) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.83 Å | |||||||||
 Authors Authors | Chang L / Wang S | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Molecular mechanism for Tn7-like transposon recruitment by a type I-B CRISPR effector. Authors: Shukun Wang / Clinton Gabel / Romana Siddique / Thomas Klose / Leifu Chang /  Abstract: Tn7-like transposons have co-opted CRISPR-Cas systems to facilitate the movement of their own DNA. These CRISPR-associated transposons (CASTs) are promising tools for programmable gene knockin. A key ...Tn7-like transposons have co-opted CRISPR-Cas systems to facilitate the movement of their own DNA. These CRISPR-associated transposons (CASTs) are promising tools for programmable gene knockin. A key feature of CASTs is their ability to recruit Tn7-like transposons to nuclease-deficient CRISPR effectors. However, how Tn7-like transposons are recruited by diverse CRISPR effectors remains poorly understood. Here, we present the cryo-EM structure of a recruitment complex comprising the Cascade complex, TniQ, TnsC, and the target DNA in the type I-B CAST from Peltigera membranacea cyanobiont 210A. Target DNA recognition by Cascade induces conformational changes in Cas6 and primes TniQ recruitment through its C-terminal domain. The N-terminal domain of TniQ is bound to the seam region of the TnsC spiral heptamer. Our findings provide insights into the diverse mechanisms for the recruitment of Tn7-like transposons to CRISPR effectors and will aid in the development of CASTs as gene knockin tools. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28980.map.gz emd_28980.map.gz | 103.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28980-v30.xml emd-28980-v30.xml emd-28980.xml emd-28980.xml | 21.2 KB 21.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28980.png emd_28980.png | 122.7 KB | ||
| Filedesc metadata |  emd-28980.cif.gz emd-28980.cif.gz | 6.5 KB | ||
| Others |  emd_28980_half_map_1.map.gz emd_28980_half_map_1.map.gz emd_28980_half_map_2.map.gz emd_28980_half_map_2.map.gz | 200.2 MB 200.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28980 http://ftp.pdbj.org/pub/emdb/structures/EMD-28980 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28980 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28980 | HTTPS FTP |
-Validation report
| Summary document |  emd_28980_validation.pdf.gz emd_28980_validation.pdf.gz | 951.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28980_full_validation.pdf.gz emd_28980_full_validation.pdf.gz | 951.2 KB | Display | |
| Data in XML |  emd_28980_validation.xml.gz emd_28980_validation.xml.gz | 16.1 KB | Display | |
| Data in CIF |  emd_28980_validation.cif.gz emd_28980_validation.cif.gz | 19 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28980 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28980 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28980 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28980 | HTTPS FTP |
-Related structure data
| Related structure data |  8fcjMC  8fcuC  8fcvC  8fcwC  8fcxC  8fd2C  8fd3C  8ff4C  8ff5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28980.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28980.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.054 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_28980_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
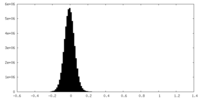
| Density Histograms |
-Half map: #1
| File | emd_28980_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of Cascade proteins, crRNA and target DNA with 23...
| Entire | Name: Ternary complex of Cascade proteins, crRNA and target DNA with 23 bp recognized. |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of Cascade proteins, crRNA and target DNA with 23...
| Supramolecule | Name: Ternary complex of Cascade proteins, crRNA and target DNA with 23 bp recognized. type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Peltigera membranacea (fungus) Peltigera membranacea (fungus) |
-Macromolecule #1: Type I-B CRISPR-associated protein Cas5
| Macromolecule | Name: Type I-B CRISPR-associated protein Cas5 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nostoc sp. 'Peltigera membranacea cyanobiont' 210A (bacteria) Nostoc sp. 'Peltigera membranacea cyanobiont' 210A (bacteria) |
| Molecular weight | Theoretical: 24.852906 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIAPLILYLD VPFTTFRESH AREMGKTYPV PPPATVYGML LSLVGETNVY RHCGVELAIA MLSSPKKSRI LRQMRRFKNA DFSHPENVI PCYQEILSNL KCLIWVRSDE EKIQPSLRER IQLAFDHPEL VRRFGCLFLG ESDQLIKTIK LAREDYLEGV R QWAIRDNR ...String: MIAPLILYLD VPFTTFRESH AREMGKTYPV PPPATVYGML LSLVGETNVY RHCGVELAIA MLSSPKKSRI LRQMRRFKNA DFSHPENVI PCYQEILSNL KCLIWVRSDE EKIQPSLRER IQLAFDHPEL VRRFGCLFLG ESDQLIKTIK LAREDYLEGV R QWAIRDNR GRLTLPYWVD HVGSRNTRFL RYRIEEMDRL SPPDLAWTMV QSPI UniProtKB: Type I-MYXAN CRISPR-associated protein Cas5/Cmx5/DevS |
-Macromolecule #2: Type I-B CRISPR-associated protein Cas6
| Macromolecule | Name: Type I-B CRISPR-associated protein Cas6 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nostoc sp. 'Peltigera membranacea cyanobiont' 210A (bacteria) Nostoc sp. 'Peltigera membranacea cyanobiont' 210A (bacteria) |
| Molecular weight | Theoretical: 24.945744 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTAILIQSEE YVDLTFKLRG APIPLDNGYL TYAALSRICP PLHELKSIGI HPIAGIPTRN NLLELTAQSR LKIRIYHQQI PLIYPYLAG QAFHIGQNFY QLDIPDYKPL ISSESVYSRL VIIKGFQDST NFIEAVQRQM DNLGIQGKIE LLTRQDGTPQ R RQLTINKE ...String: MTAILIQSEE YVDLTFKLRG APIPLDNGYL TYAALSRICP PLHELKSIGI HPIAGIPTRN NLLELTAQSR LKIRIYHQQI PLIYPYLAG QAFHIGQNFY QLDIPDYKPL ISSESVYSRL VIIKGFQDST NFIEAVQRQM DNLGIQGKIE LLTRQDGTPQ R RQLTINKE GKQFKVRGFG VKISELNPED SLTLQEQGIG GKRKMMCGIF VPATRSKEEE ET UniProtKB: Type I-MYXAN CRISPR-associated protein Cas6/Cmx6 |
-Macromolecule #3: Type I-B CRISPR-associated protein Cas7
| Macromolecule | Name: Type I-B CRISPR-associated protein Cas7 / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nostoc sp. 'Peltigera membranacea cyanobiont' 210A (bacteria) Nostoc sp. 'Peltigera membranacea cyanobiont' 210A (bacteria) |
| Molecular weight | Theoretical: 37.298996 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MMTQKKNDSN IPNYYLYGTV LTRYGLASLN HDIRRGNKTI LQKGYWNNGK IHSFVGSSAI RWALRFYLQK QGYLVNRVWD EEEHINRLT SEDFDPEKFY DDDIFGFALL ESAETEEDTS TTKRKKKQTK TSTPNQRMGA LGMNMAVSLT PYDGAVKLGA K SGREKDST ...String: MMTQKKNDSN IPNYYLYGTV LTRYGLASLN HDIRRGNKTI LQKGYWNNGK IHSFVGSSAI RWALRFYLQK QGYLVNRVWD EEEHINRLT SEDFDPEKFY DDDIFGFALL ESAETEEDTS TTKRKKKQTK TSTPNQRMGA LGMNMAVSLT PYDGAVKLGA K SGREKDST SLHFTEYHAT RYQYYFGIDA THLKDFSRIL PMIDGIMNLP KVGGSSNIFN YPFCPDSLVF QWTNHFASYI SY CFEYCDP KSKEAKLSQE FIDEVECGQI DPSKLWIGGT IVKDLQQLDN FESSPLNKAH IYRNRNEMIE ALKTVIKRDL GLE ESK UniProtKB: CRISPR-associated protein |
-Macromolecule #4: Type I-B CRISPR-associated protein Cas8
| Macromolecule | Name: Type I-B CRISPR-associated protein Cas8 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nostoc sp. 'Peltigera membranacea cyanobiont' 210A (bacteria) Nostoc sp. 'Peltigera membranacea cyanobiont' 210A (bacteria) |
| Molecular weight | Theoretical: 63.474328 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHI VSTQPKISLS LHAADTTIMH RVGMTGLYMT LKRLEKQYPL SRQRGGHISW FLTADTIELF WEGSDFIALS WLINESFQL DDTGLIHLVG LDNDRIDLRQ KIHIHEGICG VFLRLNKFYQ AGEIINTELR FEEKQVEYQY KSLTWYAHQT F AEKLCEAD ...String: MHHHHHHHHI VSTQPKISLS LHAADTTIMH RVGMTGLYMT LKRLEKQYPL SRQRGGHISW FLTADTIELF WEGSDFIALS WLINESFQL DDTGLIHLVG LDNDRIDLRQ KIHIHEGICG VFLRLNKFYQ AGEIINTELR FEEKQVEYQY KSLTWYAHQT F AEKLCEAD TQQLRHDYIQ ITSWLYLGGI VRHARTQNTT KLEEKPEYAL ALLFVPVVCH YCLLHIPSED LKERKPHRYL VV IPEIKDF EDASQRRWRL QQLETKQFHV SSLGEAGLLY YSLDDIQPEV AYYQACQVWL YEKTNKASRQ RTLMSIEEIK IDK NILITY QQVQKYFKTN YQIIKYKQIF IKVNPIRSLI ADNLVKGIHW WSNFWEKLVI EDSKEYLFNQ LFSNREGFII MAEN SEEDK QYLIFIKVFQ QAMKGNFAKI YAKTEEGKDP PIKKKVERLR AELNYCYDEL SFKEYLSDFL VRGGLNKYFN EHQEE IALL IKKSPWQEIR IWSLLAIASY KPKDKLTNRD DSSLSNNQKL EEVNDDSEEE UniProtKB: Type I-MYXAN CRISPR-associated Cas8a1/Cmx1 |
-Macromolecule #5: Type I-B CRISPR-associated protein Cas11
| Macromolecule | Name: Type I-B CRISPR-associated protein Cas11 / type: protein_or_peptide / ID: 5 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nostoc sp. 'Peltigera membranacea cyanobiont' 210A (bacteria) Nostoc sp. 'Peltigera membranacea cyanobiont' 210A (bacteria) |
| Molecular weight | Theoretical: 16.314365 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAENSEEDKQ YLIFIKVFQQ AMKGNFAKIY AKTEEGKDPP IKKKVERLRA ELNYCYDELS FKEYLSDFLV RGGLNKYFNE HQEEIALLI KKSPWQEIRI WSLLAIASYK PKDKLTNRDD SSLSNNQKLE EVNDDSEEE UniProtKB: Type I-MYXAN CRISPR-associated Cas8a1/Cmx1 |
-Macromolecule #6: RNA
| Macromolecule | Name: RNA / type: rna / ID: 6 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 22.876527 KDa |
| Sequence | String: UUGCUCAAGA GAAGUCAUUU AAUAAGGCCA CUGUUAAACG UAGGUGAGUC GUGGCUUUAU GCCGUUAGGC G |
-Macromolecule #7: Target DNA strand
| Macromolecule | Name: Target DNA strand / type: dna / ID: 7 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 19.913799 KDa |
| Sequence | String: (DA)(DT)(DA)(DT)(DC)(DT)(DA)(DC)(DG)(DC) (DG)(DT)(DA)(DG)(DA)(DT)(DA)(DT)(DA)(DT) (DC)(DT)(DA)(DC)(DG)(DT)(DT)(DT)(DA) (DA)(DC)(DA)(DG)(DT)(DG)(DG)(DC)(DC)(DT) (DT) (DA)(DT)(DT)(DA)(DA)(DA) ...String: (DA)(DT)(DA)(DT)(DC)(DT)(DA)(DC)(DG)(DC) (DG)(DT)(DA)(DG)(DA)(DT)(DA)(DT)(DA)(DT) (DC)(DT)(DA)(DC)(DG)(DT)(DT)(DT)(DA) (DA)(DC)(DA)(DG)(DT)(DG)(DG)(DC)(DC)(DT) (DT) (DA)(DT)(DT)(DA)(DA)(DA)(DT)(DG) (DA)(DC)(DT)(DT)(DC)(DT)(DC)(DC)(DA)(DT) (DG)(DA) (DT)(DC)(DT)(DA)(DC) |
-Macromolecule #8: Non-target DNA strand
| Macromolecule | Name: Non-target DNA strand / type: dna / ID: 8 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 20.149975 KDa |
| Sequence | String: (DG)(DT)(DA)(DG)(DA)(DT)(DC)(DA)(DT)(DG) (DG)(DA)(DG)(DA)(DA)(DG)(DT)(DC)(DA)(DT) (DT)(DT)(DA)(DA)(DT)(DA)(DA)(DG)(DG) (DC)(DC)(DA)(DC)(DT)(DG)(DT)(DT)(DA)(DA) (DA) (DC)(DG)(DT)(DA)(DG)(DA) ...String: (DG)(DT)(DA)(DG)(DA)(DT)(DC)(DA)(DT)(DG) (DG)(DA)(DG)(DA)(DA)(DG)(DT)(DC)(DA)(DT) (DT)(DT)(DA)(DA)(DT)(DA)(DA)(DG)(DG) (DC)(DC)(DA)(DC)(DT)(DG)(DT)(DT)(DA)(DA) (DA) (DC)(DG)(DT)(DA)(DG)(DA)(DT)(DA) (DT)(DA)(DT)(DC)(DT)(DA)(DC)(DG)(DC)(DG) (DT)(DA) (DG)(DA)(DT)(DA)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.83 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 200976 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller


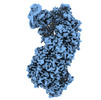









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)