[English] 日本語
 Yorodumi
Yorodumi- EMDB-28955: Asymmetric structure of cleaved HIV-1 AE2 envelope glycoprotein t... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
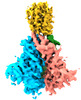
| Title | Asymmetric structure of cleaved HIV-1 AE2 envelope glycoprotein trimer in styrene-maleic acid lipid nanoparticles (AE2.2) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV-1 / envelope glycoprotein / VIRAL PROTEIN | |||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.0 Å | |||||||||
 Authors Authors | Wang K / Zhang S / Sodroski J / Mao Y | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: Asymmetric conformations of cleaved HIV-1 envelope glycoprotein trimers in styrene-maleic acid lipid nanoparticles. Authors: Kunyu Wang / Shijian Zhang / Eden P Go / Haitao Ding / Wei Li Wang / Hanh T Nguyen / John C Kappes / Heather Desaire / Joseph Sodroski / Youdong Mao /   Abstract: During virus entry, the pretriggered human immunodeficiency virus (HIV-1) envelope glycoprotein (Env) trimer initially transits into a default intermediate state (DIS) that remains structurally ...During virus entry, the pretriggered human immunodeficiency virus (HIV-1) envelope glycoprotein (Env) trimer initially transits into a default intermediate state (DIS) that remains structurally uncharacterized. Here, we present cryo-EM structures at near-atomic resolution of two cleaved full-length HIV-1 Env trimers purified from cell membranes in styrene-maleic acid lipid nanoparticles without antibodies or receptors. The cleaved Env trimers exhibited tighter subunit packing than uncleaved trimers. Cleaved and uncleaved Env trimers assumed remarkably consistent yet distinct asymmetric conformations, with one smaller and two larger opening angles. Breaking conformational symmetry is allosterically coupled with dynamic helical transformations of the gp41 N-terminal heptad repeat (HR1) regions in two protomers and with trimer tilting in the membrane. The broken symmetry of the DIS potentially assists Env binding to two CD4 receptors-while resisting antibody binding-and promotes extension of the gp41 HR1 helical coiled-coil, which relocates the fusion peptide closer to the target cell membrane. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28955.map.gz emd_28955.map.gz | 14.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28955-v30.xml emd-28955-v30.xml emd-28955.xml emd-28955.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28955.png emd_28955.png | 107.2 KB | ||
| Others |  emd_28955_half_map_1.map.gz emd_28955_half_map_1.map.gz emd_28955_half_map_2.map.gz emd_28955_half_map_2.map.gz | 11.8 MB 11.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28955 http://ftp.pdbj.org/pub/emdb/structures/EMD-28955 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28955 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28955 | HTTPS FTP |
-Validation report
| Summary document |  emd_28955_validation.pdf.gz emd_28955_validation.pdf.gz | 845.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28955_full_validation.pdf.gz emd_28955_full_validation.pdf.gz | 845.5 KB | Display | |
| Data in XML |  emd_28955_validation.xml.gz emd_28955_validation.xml.gz | 9.5 KB | Display | |
| Data in CIF |  emd_28955_validation.cif.gz emd_28955_validation.cif.gz | 11 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28955 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28955 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28955 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28955 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28955.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28955.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.65 Å | ||||||||||||||||||||||||||||||||||||
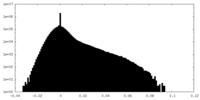
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_28955_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
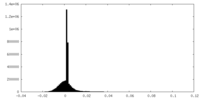
| Density Histograms |
-Half map: #2
| File | emd_28955_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
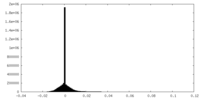
| Density Histograms |
- Sample components
Sample components
-Entire : HIV-1 Env AE2.2
| Entire | Name: HIV-1 Env AE2.2 |
|---|---|
| Components |
|
-Supramolecule #1: HIV-1 Env AE2.2
| Supramolecule | Name: HIV-1 Env AE2.2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: HIV-1 Envelop Glycoprotein
| Macromolecule | Name: HIV-1 Envelop Glycoprotein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Sequence | String: MRVKEKYQHL WRWGWRWGTM LLGMLMICSA TEKLWVTVYY GVPVWKEATT TLFCASDAKA YDTEVHNVWA THACVPTDPN PQEVVLENVT ENFNMWKNNM VEQMHEDIIS LWDESLKPCV KLTPLCVTLN CTDLRNVTNI NNSSEGMRGE IKNCSFNITT SIKDKVKKDY ...String: MRVKEKYQHL WRWGWRWGTM LLGMLMICSA TEKLWVTVYY GVPVWKEATT TLFCASDAKA YDTEVHNVWA THACVPTDPN PQEVVLENVT ENFNMWKNNM VEQMHEDIIS LWDESLKPCV KLTPLCVTLN CTDLRNVTNI NNSSEGMRGE IKNCSFNITT SIKDKVKKDY ALFYKLDVVP IDNDNTSYRL INCNTSTITQ ACPKVSFEPI PIHYCTPAGF AILKCKDKKF NGTGPCKNVS TVQCTHGIKP VVSTQLLLNG SLAEEEVVIR SSNFTDNAKN IIVQLKESVE INCTRPNNNT RKSIHIGPGK AFYTTGDIIG DIRQAHCNIS RTKWNNTLNQ IATKLKEQFG NNKTIVFNQS SGGDPEIVMH SFNCGGEFFY CNSTQLFNST WNFNGTWNLT QSNGTEGNDT ITLPCKIKQI INMWQEVGKA MYAPPIRGQI RCSSNITGLI LTRDGGNNHN NDTETFRPGG GDMRDNWRSE LYKYKVVKIE PLGVAPTKAK RRVVQREKRA VGTIGAMFLG FLGAAGSTMG AASITLTVQA RLLLSGIVQQ QNNLLKAIEA QQHLLKLTVW GIKQLQARVL TVERYLRDQQ LLGIWGCSGK LICTTAVPWN ASWSNKTLDM IWNNMTWMEW EKEIDNYTGL IYTLIEESQN QQEKNEKELL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 57.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 5.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 21068 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)