+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human CCC complex | ||||||||||||||||||||||||||||||
 Map data Map data | Sharpened refined map from cryoSPARC | ||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||
 Keywords Keywords | endosomal sorting / commander / CCC complex / ENDOCYTOSIS | ||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of sodium ion transmembrane transport / plasma membrane to endosome transport / regulation of proteasomal ubiquitin-dependent protein catabolic process / negative regulation of protein localization to cell surface / copper ion homeostasis / Golgi to plasma membrane transport / phosphatidic acid binding / sodium channel inhibitor activity / phosphatidylinositol-3,4-bisphosphate binding / phosphatidylinositol-3,5-bisphosphate binding ...negative regulation of sodium ion transmembrane transport / plasma membrane to endosome transport / regulation of proteasomal ubiquitin-dependent protein catabolic process / negative regulation of protein localization to cell surface / copper ion homeostasis / Golgi to plasma membrane transport / phosphatidic acid binding / sodium channel inhibitor activity / phosphatidylinositol-3,4-bisphosphate binding / phosphatidylinositol-3,5-bisphosphate binding / Cul2-RING ubiquitin ligase complex / negative regulation of NF-kappaB transcription factor activity / sodium ion transport / phosphatidylinositol-3,4,5-trisphosphate binding / NF-kappaB binding / phosphatidylinositol-4,5-bisphosphate binding / tumor necrosis factor-mediated signaling pathway / cholesterol homeostasis / positive regulation of protein ubiquitination / nucleotide-excision repair / recycling endosome / protein transport / Neddylation / cytoplasmic vesicle / secretory granule lumen / ficolin-1-rich granule lumen / early endosome / endosome membrane / endosome / copper ion binding / intracellular membrane-bounded organelle / negative regulation of DNA-templated transcription / Neutrophil degranulation / Golgi apparatus / protein homodimerization activity / extracellular region / nucleoplasm / identical protein binding / nucleus / plasma membrane / cytosol / cytoplasm Similarity search - Function | ||||||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||||||||
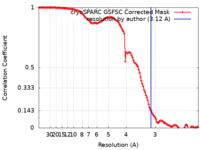
| Method | single particle reconstruction / cryo EM / Resolution: 3.12 Å | ||||||||||||||||||||||||||||||
 Authors Authors | Healy MD / McNally KE / Butkovic R / Chilton M / Kato K / Sacharz J / McConville C / Moody ERR / Shaw S / Planelles-Herrero VJ ...Healy MD / McNally KE / Butkovic R / Chilton M / Kato K / Sacharz J / McConville C / Moody ERR / Shaw S / Planelles-Herrero VJ / Kadapalakere SY / Ross J / Borucu U / Palmer CS / Chen K / Croll TI / Hall RJ / Caruana NJ / Ghai R / Nguyen THD / Heesom KJ / Saitoh S / Berger I / Berger-Schaffitzel C / Williams TA / Stroud DA / Derivery E / Collins BM / Cullen PJ | ||||||||||||||||||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  Australia, 9 items Australia, 9 items
| ||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Structure of the endosomal Commander complex linked to Ritscher-Schinzel syndrome. Authors: Michael D Healy / Kerrie E McNally / Rebeka Butkovič / Molly Chilton / Kohji Kato / Joanna Sacharz / Calum McConville / Edmund R R Moody / Shrestha Shaw / Vicente J Planelles-Herrero / ...Authors: Michael D Healy / Kerrie E McNally / Rebeka Butkovič / Molly Chilton / Kohji Kato / Joanna Sacharz / Calum McConville / Edmund R R Moody / Shrestha Shaw / Vicente J Planelles-Herrero / Sathish K N Yadav / Jennifer Ross / Ufuk Borucu / Catherine S Palmer / Kai-En Chen / Tristan I Croll / Ryan J Hall / Nikeisha J Caruana / Rajesh Ghai / Thi H D Nguyen / Kate J Heesom / Shinji Saitoh / Imre Berger / Christiane Schaffitzel / Tom A Williams / David A Stroud / Emmanuel Derivery / Brett M Collins / Peter J Cullen /    Abstract: The Commander complex is required for endosomal recycling of diverse transmembrane cargos and is mutated in Ritscher-Schinzel syndrome. It comprises two sub-assemblies: Retriever composed of VPS35L, ...The Commander complex is required for endosomal recycling of diverse transmembrane cargos and is mutated in Ritscher-Schinzel syndrome. It comprises two sub-assemblies: Retriever composed of VPS35L, VPS26C, and VPS29; and the CCC complex which contains twelve subunits: COMMD1-COMMD10 and the coiled-coil domain-containing (CCDC) proteins CCDC22 and CCDC93. Combining X-ray crystallography, electron cryomicroscopy, and in silico predictions, we have assembled a complete structural model of Commander. Retriever is distantly related to the endosomal Retromer complex but has unique features preventing the shared VPS29 subunit from interacting with Retromer-associated factors. The COMMD proteins form a distinctive hetero-decameric ring stabilized by extensive interactions with CCDC22 and CCDC93. These adopt a coiled-coil structure that connects the CCC and Retriever assemblies and recruits a 16th subunit, DENND10, to form the complete Commander complex. The structure allows mapping of disease-causing mutations and reveals the molecular features required for the function of this evolutionarily conserved trafficking machinery. | ||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28825.map.gz emd_28825.map.gz | 168.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28825-v30.xml emd-28825-v30.xml emd-28825.xml emd-28825.xml | 32 KB 32 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28825_fsc.xml emd_28825_fsc.xml | 11.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_28825.png emd_28825.png | 51.7 KB | ||
| Filedesc metadata |  emd-28825.cif.gz emd-28825.cif.gz | 7.7 KB | ||
| Others |  emd_28825_additional_1.map.gz emd_28825_additional_1.map.gz emd_28825_half_map_1.map.gz emd_28825_half_map_1.map.gz emd_28825_half_map_2.map.gz emd_28825_half_map_2.map.gz | 89.6 MB 165.4 MB 165.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28825 http://ftp.pdbj.org/pub/emdb/structures/EMD-28825 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28825 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28825 | HTTPS FTP |
-Validation report
| Summary document |  emd_28825_validation.pdf.gz emd_28825_validation.pdf.gz | 880.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28825_full_validation.pdf.gz emd_28825_full_validation.pdf.gz | 879.8 KB | Display | |
| Data in XML |  emd_28825_validation.xml.gz emd_28825_validation.xml.gz | 20.5 KB | Display | |
| Data in CIF |  emd_28825_validation.cif.gz emd_28825_validation.cif.gz | 26.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28825 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28825 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28825 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28825 | HTTPS FTP |
-Related structure data
| Related structure data |  8f2rMC  8esdC  8eseC  8f2uC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28825.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28825.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened refined map from cryoSPARC | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.084 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened refined map
| File | emd_28825_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened refined map | ||||||||||||
| Projections & Slices |
| ||||||||||||
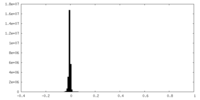
| Density Histograms |
-Half map: Half map
| File | emd_28825_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
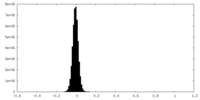
| Density Histograms |
-Half map: Half map
| File | emd_28825_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
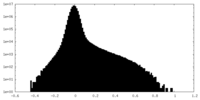
| Density Histograms |
- Sample components
Sample components
+Entire : Human CCC complex
+Supramolecule #1: Human CCC complex
+Macromolecule #1: COMM domain-containing protein 1
+Macromolecule #2: COMM domain-containing protein 2
+Macromolecule #3: COMM domain-containing protein 3
+Macromolecule #4: COMM domain-containing protein 4
+Macromolecule #5: COMM domain-containing protein 5
+Macromolecule #6: COMM domain-containing protein 6
+Macromolecule #7: COMM domain-containing protein 7
+Macromolecule #8: COMM domain-containing protein 8
+Macromolecule #9: COMM domain-containing protein 9
+Macromolecule #10: COMM domain-containing protein 10
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.2 Details: 50 mM HEPES pH7.2, 150 mM NaCl, 2mM beta-mercaptoethanol, 0.01% Triton-X100 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: GRAPHENE OXIDE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
| Details | Human CCC complex cross-linked with 1 mM BS3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 2 / Number real images: 5651 / Average exposure time: 9.01 sec. / Average electron dose: 43.14 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)








































 Spodoptera (butterflies/moths)
Spodoptera (butterflies/moths)