+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Local refinement of P8 domain of LRP2 at pH 5.2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Beenken A / Shapiro L | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Structures of LRP2 reveal a molecular machine for endocytosis. Authors: Andrew Beenken / Gabriele Cerutti / Julia Brasch / Yicheng Guo / Zizhang Sheng / Hediye Erdjument-Bromage / Zainab Aziz / Shelief Y Robbins-Juarez / Estefania Y Chavez / Goran Ahlsen / ...Authors: Andrew Beenken / Gabriele Cerutti / Julia Brasch / Yicheng Guo / Zizhang Sheng / Hediye Erdjument-Bromage / Zainab Aziz / Shelief Y Robbins-Juarez / Estefania Y Chavez / Goran Ahlsen / Phinikoula S Katsamba / Thomas A Neubert / Anthony W P Fitzpatrick / Jonathan Barasch / Lawrence Shapiro /  Abstract: The low-density lipoprotein (LDL) receptor-related protein 2 (LRP2 or megalin) is representative of the phylogenetically conserved subfamily of giant LDL receptor-related proteins, which function in ...The low-density lipoprotein (LDL) receptor-related protein 2 (LRP2 or megalin) is representative of the phylogenetically conserved subfamily of giant LDL receptor-related proteins, which function in endocytosis and are implicated in diseases of the kidney and brain. Here, we report high-resolution cryoelectron microscopy structures of LRP2 isolated from mouse kidney, at extracellular and endosomal pH. The structures reveal LRP2 to be a molecular machine that adopts a conformation for ligand binding at the cell surface and for ligand shedding in the endosome. LRP2 forms a homodimer, the conformational transformation of which is governed by pH-sensitive sites at both homodimer and intra-protomer interfaces. A subset of LRP2 deleterious missense variants in humans appears to impair homodimer assembly. These observations lay the foundation for further understanding the function and mechanism of LDL receptors and implicate homodimerization as a conserved feature of the LRP receptor subfamily. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28265.map.gz emd_28265.map.gz | 256.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28265-v30.xml emd-28265-v30.xml emd-28265.xml emd-28265.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28265_fsc.xml emd_28265_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_28265.png emd_28265.png | 84.8 KB | ||
| Others |  emd_28265_additional_1.map.gz emd_28265_additional_1.map.gz emd_28265_half_map_1.map.gz emd_28265_half_map_1.map.gz emd_28265_half_map_2.map.gz emd_28265_half_map_2.map.gz | 424.7 MB 475.3 MB 475.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28265 http://ftp.pdbj.org/pub/emdb/structures/EMD-28265 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28265 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28265 | HTTPS FTP |
-Validation report
| Summary document |  emd_28265_validation.pdf.gz emd_28265_validation.pdf.gz | 875.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28265_full_validation.pdf.gz emd_28265_full_validation.pdf.gz | 874.9 KB | Display | |
| Data in XML |  emd_28265_validation.xml.gz emd_28265_validation.xml.gz | 26.1 KB | Display | |
| Data in CIF |  emd_28265_validation.cif.gz emd_28265_validation.cif.gz | 34.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28265 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28265 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28265 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28265 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28265.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28265.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
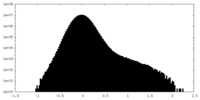
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_28265_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
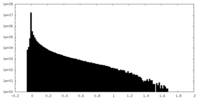
| Density Histograms |
-Half map: #1
| File | emd_28265_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_28265_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : LRP2 at endosomal pH
| Entire | Name: LRP2 at endosomal pH |
|---|---|
| Components |
|
-Supramolecule #1: LRP2 at endosomal pH
| Supramolecule | Name: LRP2 at endosomal pH / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Details: Endogenously purified from mouse kidney |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 5.2 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 58.06 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)