[English] 日本語
 Yorodumi
Yorodumi- EMDB-27426: Avs4 bound to phage PhiV-1 portal, symmetry-expanded C1 refinemen... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Avs4 bound to phage PhiV-1 portal, symmetry-expanded C1 refinement of TPR-portal domain | ||||||||||||
 Map data Map data | Avs4 bound to phage PhiV-1 portal, symmetry-expanded C1 refinement of TPR-portal domain | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |   Escherichia phage PhiV-1 (virus) Escherichia phage PhiV-1 (virus) | ||||||||||||
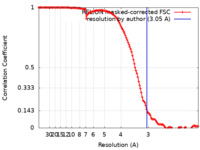
| Method | single particle reconstruction / cryo EM / Resolution: 3.05 Å | ||||||||||||
 Authors Authors | Wilkinson ME / Gao L / Strecker J / Makarova KS / Macrae RK / Koonin EV / Zhang F | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Prokaryotic innate immunity through pattern recognition of conserved viral proteins. Authors: Linyi Alex Gao / Max E Wilkinson / Jonathan Strecker / Kira S Makarova / Rhiannon K Macrae / Eugene V Koonin / Feng Zhang /  Abstract: Many organisms have evolved specialized immune pattern-recognition receptors, including nucleotide-binding oligomerization domain-like receptors (NLRs) of the STAND superfamily that are ubiquitous in ...Many organisms have evolved specialized immune pattern-recognition receptors, including nucleotide-binding oligomerization domain-like receptors (NLRs) of the STAND superfamily that are ubiquitous in plants, animals, and fungi. Although the roles of NLRs in eukaryotic immunity are well established, it is unknown whether prokaryotes use similar defense mechanisms. Here, we show that antiviral STAND (Avs) homologs in bacteria and archaea detect hallmark viral proteins, triggering Avs tetramerization and the activation of diverse N-terminal effector domains, including DNA endonucleases, to abrogate infection. Cryo-electron microscopy reveals that Avs sensor domains recognize conserved folds, active-site residues, and enzyme ligands, allowing a single Avs receptor to detect a wide variety of viruses. These findings extend the paradigm of pattern recognition of pathogen-specific proteins across all three domains of life. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27426.map.gz emd_27426.map.gz | 162.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27426-v30.xml emd-27426-v30.xml emd-27426.xml emd-27426.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27426_fsc.xml emd_27426_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_27426.png emd_27426.png | 57.7 KB | ||
| Masks |  emd_27426_msk_1.map emd_27426_msk_1.map | 178 MB |  Mask map Mask map | |
| Others |  emd_27426_half_map_1.map.gz emd_27426_half_map_1.map.gz emd_27426_half_map_2.map.gz emd_27426_half_map_2.map.gz | 140.6 MB 140.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27426 http://ftp.pdbj.org/pub/emdb/structures/EMD-27426 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27426 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27426 | HTTPS FTP |
-Validation report
| Summary document |  emd_27426_validation.pdf.gz emd_27426_validation.pdf.gz | 838.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27426_full_validation.pdf.gz emd_27426_full_validation.pdf.gz | 838.1 KB | Display | |
| Data in XML |  emd_27426_validation.xml.gz emd_27426_validation.xml.gz | 20 KB | Display | |
| Data in CIF |  emd_27426_validation.cif.gz emd_27426_validation.cif.gz | 26.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27426 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27426 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27426 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27426 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27426.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27426.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Avs4 bound to phage PhiV-1 portal, symmetry-expanded C1 refinement of TPR-portal domain | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0344 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27426_msk_1.map emd_27426_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_27426_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_27426_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Avs4 bound to phage PhiV-1 portal
| Entire | Name: Avs4 bound to phage PhiV-1 portal |
|---|---|
| Components |
|
-Supramolecule #1: Avs4 bound to phage PhiV-1 portal
| Supramolecule | Name: Avs4 bound to phage PhiV-1 portal / type: complex / Chimera: Yes / ID: 1 / Parent: 0 |
|---|---|
| Molecular weight | Theoretical: 980 KDa |
-Supramolecule #2: Avs4
| Supramolecule | Name: Avs4 / type: complex / Chimera: Yes / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Supramolecule #3: phage PhiV-1 portal
| Supramolecule | Name: phage PhiV-1 portal / type: complex / Chimera: Yes / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Escherichia phage PhiV-1 (virus) Escherichia phage PhiV-1 (virus) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.7 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 31.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)