[English] 日本語
 Yorodumi
Yorodumi- EMDB-27422: Avs4 bound to phage PhiV-1 portal, C2 refinement of Mrr nuclease ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Avs4 bound to phage PhiV-1 portal, C2 refinement of Mrr nuclease domain | ||||||||||||
 Map data Map data | Avs4 bound to phage PhiV-1 portal, C2 refinement of Mrr nuclease domain | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | phage defense / pattern-recognition receptor / nlr / stand / atpase / ANTIVIRAL PROTEIN | ||||||||||||
| Function / homology | Portal protein, Caudovirales / Head-to-tail connector protein, podovirus-type / Bacteriophage head to tail connecting protein / viral portal complex / symbiont genome ejection through host cell envelope, short tail mechanism / viral DNA genome packaging / Portal protein Function and homology information Function and homology information | ||||||||||||
| Biological species |   Escherichia phage PhiV-1 (virus) Escherichia phage PhiV-1 (virus) | ||||||||||||
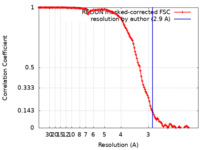
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | ||||||||||||
 Authors Authors | Wilkinson ME / Gao L / Strecker J / Makarova KS / Macrae RK / Koonin EV / Zhang F | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Prokaryotic innate immunity through pattern recognition of conserved viral proteins. Authors: Linyi Alex Gao / Max E Wilkinson / Jonathan Strecker / Kira S Makarova / Rhiannon K Macrae / Eugene V Koonin / Feng Zhang /  Abstract: Many organisms have evolved specialized immune pattern-recognition receptors, including nucleotide-binding oligomerization domain-like receptors (NLRs) of the STAND superfamily that are ubiquitous in ...Many organisms have evolved specialized immune pattern-recognition receptors, including nucleotide-binding oligomerization domain-like receptors (NLRs) of the STAND superfamily that are ubiquitous in plants, animals, and fungi. Although the roles of NLRs in eukaryotic immunity are well established, it is unknown whether prokaryotes use similar defense mechanisms. Here, we show that antiviral STAND (Avs) homologs in bacteria and archaea detect hallmark viral proteins, triggering Avs tetramerization and the activation of diverse N-terminal effector domains, including DNA endonucleases, to abrogate infection. Cryo-electron microscopy reveals that Avs sensor domains recognize conserved folds, active-site residues, and enzyme ligands, allowing a single Avs receptor to detect a wide variety of viruses. These findings extend the paradigm of pattern recognition of pathogen-specific proteins across all three domains of life. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27422.map.gz emd_27422.map.gz | 161.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27422-v30.xml emd-27422-v30.xml emd-27422.xml emd-27422.xml | 22 KB 22 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27422_fsc.xml emd_27422_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_27422.png emd_27422.png | 55.9 KB | ||
| Masks |  emd_27422_msk_1.map emd_27422_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27422.cif.gz emd-27422.cif.gz | 7.7 KB | ||
| Others |  emd_27422_half_map_1.map.gz emd_27422_half_map_1.map.gz emd_27422_half_map_2.map.gz emd_27422_half_map_2.map.gz | 139.5 MB 139.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27422 http://ftp.pdbj.org/pub/emdb/structures/EMD-27422 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27422 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27422 | HTTPS FTP |
-Related structure data
| Related structure data |  8dgfMC  8dgcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27422.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27422.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Avs4 bound to phage PhiV-1 portal, C2 refinement of Mrr nuclease domain | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03436 Å | ||||||||||||||||||||||||||||||||||||
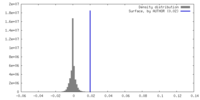
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27422_msk_1.map emd_27422_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
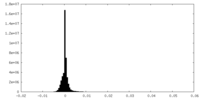
| Density Histograms |
-Half map: Half map 1
| File | emd_27422_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_27422_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Avs4 bound to phage PhiV-1 portal
| Entire | Name: Avs4 bound to phage PhiV-1 portal |
|---|---|
| Components |
|
-Supramolecule #1: Avs4 bound to phage PhiV-1 portal
| Supramolecule | Name: Avs4 bound to phage PhiV-1 portal / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Molecular weight | Theoretical: 980 KDa |
-Macromolecule #1: ATP-binding protein Avs4
| Macromolecule | Name: ATP-binding protein Avs4 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 186.640906 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: (FME)VKPNWDNFK AKFSENPQGN FEWFCYLLFC QEFKMPAGIF RYKNQSGIET NPITKDNEII GWQSKFYDTK LSDNKA DLI EMIEKSKKAY PGLSKIIFYT NQEWGQGRKS HEPEGDKNAD NYLETVGNSN DPKIKIEVDQ KAYESGIEIV WRVASFF ES PFVIVENEKI ...String: (FME)VKPNWDNFK AKFSENPQGN FEWFCYLLFC QEFKMPAGIF RYKNQSGIET NPITKDNEII GWQSKFYDTK LSDNKA DLI EMIEKSKKAY PGLSKIIFYT NQEWGQGRKS HEPEGDKNAD NYLETVGNSN DPKIKIEVDQ KAYESGIEIV WRVASFF ES PFVIVENEKI AKHFFSLNES IFDLLEEKRK HTENVLYEIQ TNIEFKDRSI EIDRRHCIEL LHENLVQKKI VIVSGEGG V GKTAVIKKIY EAEKQYTPFY VFKASEFKKD SINELFGAHG LDDFSNAHQD ELRKVIVVDS AEKLLELTNI DPFKEFLTV LIKDKWQVVF TTRNNYLADL NYAFIDIYKI TPGNLVIKNL ERGELIELSD NNGFSLPQDV RLLELIKNPF YLSEYLRFYT GESIDYVSF KEKLWNKIIV KNKPSREQCF LATAFQRASE GQFFVSPACD TGILDELVKD GIVGYEAAGY FITHDIYEEW A LEKKISVD YIRKANNNEF FEKIGESLPV RRSFRNWISE RLLLDDQSIK PFIAEIVCGE GISNFWKDEL WVAVLLSDNS SI FFNYFKR YLLSSDQNLL KRLTFLLRLA CKDVDYDLLK QLGVSNSDLL SIKYVLTKPK GTGWQSVIQF IYENLDEIGI RNI NFILPV IQEWNQRNKV GETTRLSSLI ALKYYQWTID EDVYLSGRDN EKNILHTILH GAAMIKPEME EVLVKVLKNR WKEH GTPYF DLMTLILTDL DSYPVWASLP EYVLQLADLF WYRPLKETGE RYHSMDIEDE FGLFRSHHDY YPESPYQTPI YWLLQ SQFK KTIDFILDFT NKTTICFAHS HFAKNEIEEV DVFIEEGKFI KQYICNRLWC SYRGTQVSTY LLSSIHMALE KFFLEN FKN ADSKVLESWL LFLLRNTKSA SISAVVTSIV LAFPEKTFNV AKVLFQTKDF FRFDMNRMVL DRTHKSSLIS LRDGFGG TD YRNSLHEEDR IKACDDVHRN TYLENLALHY QIFRSENVTE KDAIERQQVL WDIFDKYYNQ LPDEAQETEA DKTWRLCL A RMDRRKMKIT TKEKDEGIEI SFNPEIDPKL KQYSEEAIKK NSEHMKYVTL KLWASYKREK DERYKNYGMY EDNPQIALQ ETKEIIKKLN EEGGEDFRLL NGNIPADVCS VLLLDYFNQL NNEEREYCKD IVLAYSKLPL KEGYNYQVQD GTTSAISALP VIYHNYPME RETIKTILLL TLFNDHSIGM AGGRYSVFPS MVIHKLWLDY FDDMQSLLFG FLILKPKYVI LSRKIIHESY R QVDYDIKK ININKVFLNN YKHCISNVID NKISIDDLGS MDKVDLHILN TAFQLIPVDT VNIEHKKLVS LIVKRFSTSL LS SVREDRV DYALRQSFLE RFAYFTLHAP VSDIPDYIKP FLDGFNGSEP ISELFKKFIL VEDRLNTYAK FWKVWDLFFD KVV TLCKDG DRYWYVDKII KSYLFAESPW KENSNGWHTF KDSNSQFFCD VSRTMGHCPS TLYSLAKSLN NIASCYLNQG ITWL SEILS VNKKLWEKKL ENDTVYYLEC LVRRYINNER ERIRRTKQLK QEVLVILDFL VEKGSVVGYM SRENIL |
-Macromolecule #2: Portal protein
| Macromolecule | Name: Portal protein / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage PhiV-1 (virus) Escherichia phage PhiV-1 (virus) |
| Molecular weight | Theoretical: 58.556 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASSQKREGF AENGAKAVYD ALKNDRNSYE TRAENCAKYT IPSLFPKDSD NASTDYTTPW QAVGARGLNN LASKLMLALF PMQTWMKLT ISEFEAKQLV AQPAELAKVE EGLSMVERIL MNYIESNSYR VTLFETLKQL VVAGNALLYI PEPEGAYNPM K LYRLSSYV ...String: MASSQKREGF AENGAKAVYD ALKNDRNSYE TRAENCAKYT IPSLFPKDSD NASTDYTTPW QAVGARGLNN LASKLMLALF PMQTWMKLT ISEFEAKQLV AQPAELAKVE EGLSMVERIL MNYIESNSYR VTLFETLKQL VVAGNALLYI PEPEGAYNPM K LYRLSSYV VQRDAFGTVL QIVTLDKTAY AALPEDVRNA MDSGQEHKGD EMIDVYTHIY LDEESGEYLK YEEIDGVEVD GT DASYPVD ACPYIPVRMV RIDGESYGRS YCEEYLGDLR SLENLQEAIV KMSMISAKVI GLVNPAGITQ VRRLTKAQTG DFV SGRPED ISFLQLEKAA DFSVAKAVSE QIEGRLSYAF MLNSAVQRTG ERVTAEEIRY VASELEDTLG GVYSILSQEL QLPM VRVLL KQLQATNQIP ELPKEAVEPT ISTGMEALGR GQDLDKLERC IAAWSALAPM QNDPDINIAT IKLRIANAIG IDTSG ILKT PEEKQQEMAE AAQGTALENA AASAGAGAGA LATASPENME AAAAQAGMVP N UniProtKB: Portal protein |
-Macromolecule #3: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 4 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.7 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 31.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)