[English] 日本語
 Yorodumi
Yorodumi- EMDB-27407: Group A streptococcus Enolase K252A, K255A, K434A, K435A mutant -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Group A streptococcus Enolase K252A, K255A, K434A, K435A mutant | |||||||||
 Map data Map data | GAS enolase with K252A, K255A, K434A, and K435 mutations made full map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | metalloenzyme / hPg-receptor / LYASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationphosphopyruvate hydratase / phosphopyruvate hydratase complex / phosphopyruvate hydratase activity / peptidoglycan-based cell wall / glycolytic process / cell surface / magnesium ion binding / extracellular region Similarity search - Function | |||||||||
| Biological species |  Streptococcus pyogenes (bacteria) / Streptococcus pyogenes (bacteria) /  Streptococcus sp. 'group A' (bacteria) Streptococcus sp. 'group A' (bacteria) | |||||||||
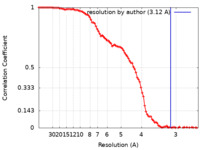
| Method | single particle reconstruction / cryo EM / Resolution: 3.12 Å | |||||||||
 Authors Authors | Tjia-Fleck SC / Readnour BM / Castellino FJ | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Biochemistry / Year: 2023 Journal: Biochemistry / Year: 2023Title: High-Resolution Single-Particle Cryo-EM Hydrated Structure of Enolase Offers Insights into Its Function as a Plasminogen Receptor. Authors: Sheiny Tjia-Fleck / Bradley M Readnour / Yetunde A Ayinuola / Francis J Castellino /  Abstract: Cellular plasminogen (Pg) receptors (PgRs) are utilized to recruit Pg; stimulate its activation to the serine protease, plasmin (Pm); and sterically protect the surface Pm from inactivation by host ...Cellular plasminogen (Pg) receptors (PgRs) are utilized to recruit Pg; stimulate its activation to the serine protease, plasmin (Pm); and sterically protect the surface Pm from inactivation by host inhibitors. One such PgR is the moonlighting enzyme, enolase, some of which leaves the cytoplasm and resides at the cell surface to potentially function as a PgR. Since microbes employ conscription of host Pg by PgRs as one virulence mechanism, we explored the structural basis of the ability of enolase (Sen) to function in this manner. Employing single-particle cryo-electron microscopy (cryo-EM), recombinant Sen from was modeled at 2.6 Å as a stable symmetrical doughnut-shaped homooctamer with point group 422 (D4) symmetry, with a monomeric subunit molecular weight of ∼49 kDa. Binding sites for hPg were reported in other studies to include an internal K and the COOH-terminal K residues of Sen. However, in native Sen, the latter are buried within the minor interfaces of the octamer and do not function as a Pg-binding epitope. Whereas Sen and hPg do not interact in solution, when Sen is bound to a surface, hPg interacts with Sen independently of K. PgRs devoid of COOH-terminal lysine utilize lysine isosteres comprising a basic residue, "", and an anionic residue at " + 3" around one turn of an α-helix. We highlight a number of surface-exposed potential hPg-binding lysine isosteres and further conclude that while the octameric structure of Sen is critical for hPg binding, disruption of this octamer without dissociation exposes hPg-binding epitopes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27407.map.gz emd_27407.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27407-v30.xml emd-27407-v30.xml emd-27407.xml emd-27407.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27407_fsc.xml emd_27407_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_27407.png emd_27407.png | 150 KB | ||
| Masks |  emd_27407_msk_1.map emd_27407_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_27407_half_map_1.map.gz emd_27407_half_map_1.map.gz emd_27407_half_map_2.map.gz emd_27407_half_map_2.map.gz | 59.1 MB 59.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27407 http://ftp.pdbj.org/pub/emdb/structures/EMD-27407 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27407 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27407 | HTTPS FTP |
-Validation report
| Summary document |  emd_27407_validation.pdf.gz emd_27407_validation.pdf.gz | 979.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27407_full_validation.pdf.gz emd_27407_full_validation.pdf.gz | 979.5 KB | Display | |
| Data in XML |  emd_27407_validation.xml.gz emd_27407_validation.xml.gz | 15.8 KB | Display | |
| Data in CIF |  emd_27407_validation.cif.gz emd_27407_validation.cif.gz | 20.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27407 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27407 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27407 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27407 | HTTPS FTP |
-Related structure data
| Related structure data |  8dg4MC  7uguC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27407.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27407.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GAS enolase with K252A, K255A, K434A, and K435 mutations made full map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.29 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27407_msk_1.map emd_27407_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: GAS enolase with K252A, K255A, K434A, and K435...
| File | emd_27407_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GAS enolase with K252A, K255A, K434A, and K435 mutations made first half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: GAS enolase with K252A, K255A, K434A, and K435...
| File | emd_27407_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GAS enolase with K252A, K255A, K434A, and K435 mutations made second half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Octameric Structure of Enolase from Streptococcus Pyogenes
| Entire | Name: Octameric Structure of Enolase from Streptococcus Pyogenes |
|---|---|
| Components |
|
-Supramolecule #1: Octameric Structure of Enolase from Streptococcus Pyogenes
| Supramolecule | Name: Octameric Structure of Enolase from Streptococcus Pyogenes type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes (bacteria) / Strain: AP53 Streptococcus pyogenes (bacteria) / Strain: AP53 |
| Molecular weight | Theoretical: 400 KDa |
-Macromolecule #1: Enolase
| Macromolecule | Name: Enolase / type: protein_or_peptide / ID: 1 / Enantiomer: DEXTRO |
|---|---|
| Source (natural) | Organism:  Streptococcus sp. 'group A' (bacteria) / Strain: AP53 Streptococcus sp. 'group A' (bacteria) / Strain: AP53 |
| Recombinant expression | Organism:  |
| Sequence | String: HMSIITDVYA REVLDSRGNP TLEVEVYTES GAFGRGMVPS GASTGEHEAV ELRDGDKSRY LGLGTQKAVD NVNNIIAEAI IGYDVRDQQA IDRAMIALDG TPNKGKLGAN AILGVSIAVA RAAADYLEVP LYTYLGGFNT KVLPTPMMNI INGGSHSDAP IAFQEFMIMP ...String: HMSIITDVYA REVLDSRGNP TLEVEVYTES GAFGRGMVPS GASTGEHEAV ELRDGDKSRY LGLGTQKAVD NVNNIIAEAI IGYDVRDQQA IDRAMIALDG TPNKGKLGAN AILGVSIAVA RAAADYLEVP LYTYLGGFNT KVLPTPMMNI INGGSHSDAP IAFQEFMIMP VGAPTFKEGL RWGAEVFHAL KKILKERGLV TAVGDEGGFA PKFEGTEDGV ETILKAIEAA GYEAGENGIM IGFDCASSEF YDAERAVYDY TKFEGEGAAV RTSAEQVDYL EELVNKYPII TIEDGMDEND WDGWKVLTER LGKRVQLVGD DFFVTNTEYL ARGIKENAAN SILIKVNQIG TLTETFEAIE MAKEAGYTAV VSHRSGETED STIADIAVAT NAGQIKTGSL SRTDRIAKYN QLLRIEDQLG EVAQYKGIKS FYNLAA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Component - Concentration: 0.05 mM / Component - Formula: NaH2PO4 / Component - Name: Sodium Phosphate |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Number grids imaged: 1 / Number real images: 2756 / Average electron dose: 61.37 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 1.1 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X