+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Intracellular Coxiella 6hpi | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Coxiella burnetii (bacteria) Coxiella burnetii (bacteria) | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Park D / Liu J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Infect Immun / Year: 2022 Journal: Infect Immun / Year: 2022Title: Developmental Transitions Coordinate Assembly of the Coxiella burnetii Dot/Icm Type IV Secretion System. Authors: Donghyun Park / Samuel Steiner / Meng Shao / Craig R Roy / Jun Liu /  Abstract: Coxiella burnetii is an obligate intracellular bacterial pathogen that has evolved a unique biphasic developmental cycle. The infectious form of C. burnetii is the dormant small cell variant (SCV), ...Coxiella burnetii is an obligate intracellular bacterial pathogen that has evolved a unique biphasic developmental cycle. The infectious form of C. burnetii is the dormant small cell variant (SCV), which transitions to a metabolically active large cell variant (LCV) that replicates inside the lysosome-derived host vacuole. A Dot/Icm type IV secretion system (T4SS), which can deliver over 100 effector proteins to host cells, is essential for the biogenesis of the vacuole and intracellular replication. How the distinct C. burnetii life cycle impacts the assembly and function of the Dot/Icm T4SS has remained unknown. Here, we combine advanced cryo-focused ion beam (cryo-FIB) milling and cryo-electron tomography (cryo-ET) imaging to visualize all developmental transitions and the assembly of the Dot/Icm T4SS . Importantly, assembled Dot/Icm machines were not present in the infectious SCV. The appearance of the assembled Dot/Icm machine correlated with the transition of the SCV to the LCV intracellularly. Furthermore, temporal characterization of C. burnetii morphological changes revealed regions of the inner membrane that invaginate to form tightly packed stacks during the LCV-to-SCV transition at late stages of infection, which may enable the SCV-to-LCV transition that occurs upon infection of a new host cell. Overall, these data establish how C. burnetii developmental transitions control critical bacterial processes to promote intracellular replication and transmission. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27111.map.gz emd_27111.map.gz | 691.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27111-v30.xml emd-27111-v30.xml emd-27111.xml emd-27111.xml | 7.9 KB 7.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27111.png emd_27111.png | 210.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27111 http://ftp.pdbj.org/pub/emdb/structures/EMD-27111 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27111 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27111 | HTTPS FTP |
-Validation report
| Summary document |  emd_27111_validation.pdf.gz emd_27111_validation.pdf.gz | 373.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27111_full_validation.pdf.gz emd_27111_full_validation.pdf.gz | 372.8 KB | Display | |
| Data in XML |  emd_27111_validation.xml.gz emd_27111_validation.xml.gz | 4.9 KB | Display | |
| Data in CIF |  emd_27111_validation.cif.gz emd_27111_validation.cif.gz | 5.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27111 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27111 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27111 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27111 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27111.map.gz / Format: CCP4 / Size: 1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27111.map.gz / Format: CCP4 / Size: 1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 18.22 Å | ||||||||||||||||||||||||||||||||
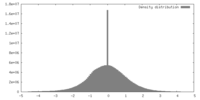
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : COS-7 cell infected with Coxiella 6hpi
| Entire | Name: COS-7 cell infected with Coxiella 6hpi |
|---|---|
| Components |
|
-Supramolecule #1: COS-7 cell infected with Coxiella 6hpi
| Supramolecule | Name: COS-7 cell infected with Coxiella 6hpi / type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Coxiella burnetii (bacteria) Coxiella burnetii (bacteria) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Sectioning | Focused ion beam - Instrument: OTHER / Focused ion beam - Ion: OTHER / Focused ion beam - Voltage: 3 kV / Focused ion beam - Current: 0.05 nA / Focused ion beam - Duration: 3600 sec. / Focused ion beam - Temperature: 103 K / Focused ion beam - Initial thickness: 10000 nm / Focused ion beam - Final thickness: 150 nm Focused ion beam - Details: The value given for _emd_sectioning_focused_ion_beam.instrument is FEI Aquilos. This is not in a list of allowed values {'OTHER', 'DB235'} so OTHER is written into the XML file. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 0.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Number images used: 33 |
|---|
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)