[English] 日本語
 Yorodumi
Yorodumi- EMDB-26984: CryoEM structure of human S-OPA1 assembled on lipid membrane in m... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of human S-OPA1 assembled on lipid membrane in membrane-distal state | |||||||||
 Map data Map data | Refine3D map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GTPase / polymer / filament / membrane / remodeling / fusion / mitochondria / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationRegulation of Apoptosis / mitochondrial inner membrane fusion / membrane tubulation / GTPase-dependent fusogenic activity / membrane bending activity / inner mitochondrial membrane organization / dynamin GTPase / cristae formation / mitochondrial genome maintenance / phosphatidic acid binding ...Regulation of Apoptosis / mitochondrial inner membrane fusion / membrane tubulation / GTPase-dependent fusogenic activity / membrane bending activity / inner mitochondrial membrane organization / dynamin GTPase / cristae formation / mitochondrial genome maintenance / phosphatidic acid binding / cardiolipin binding / mitochondrial fission / GTP metabolic process / mitochondrial fusion / axonal transport of mitochondrion / protein complex oligomerization / negative regulation of release of cytochrome c from mitochondria / mitochondrial crista / positive regulation of interleukin-17 production / negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway / positive regulation of T-helper 17 cell lineage commitment / axon cytoplasm / Mitochondrial protein degradation / visual perception / neural tube closure / mitochondrion organization / mitochondrial membrane / mitochondrial intermembrane space / cellular senescence / microtubule binding / mitochondrial outer membrane / microtubule / mitochondrial inner membrane / GTPase activity / dendrite / negative regulation of apoptotic process / GTP binding / apoptotic process / magnesium ion binding / mitochondrion / nucleoplasm / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
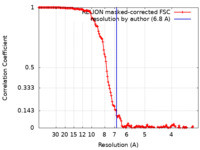
| Method | helical reconstruction / cryo EM / Resolution: 6.8 Å | |||||||||
 Authors Authors | Du Pont KE / Aydin H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Structural mechanism of mitochondrial membrane remodelling by human OPA1. Authors: Alexander von der Malsburg / Gracie M Sapp / Kelly E Zuccaro / Alexander von Appen / Frank R Moss / Raghav Kalia / Jeremy A Bennett / Luciano A Abriata / Matteo Dal Peraro / Martin van der ...Authors: Alexander von der Malsburg / Gracie M Sapp / Kelly E Zuccaro / Alexander von Appen / Frank R Moss / Raghav Kalia / Jeremy A Bennett / Luciano A Abriata / Matteo Dal Peraro / Martin van der Laan / Adam Frost / Halil Aydin /    Abstract: Distinct morphologies of the mitochondrial network support divergent metabolic and regulatory processes that determine cell function and fate. The mechanochemical GTPase optic atrophy 1 (OPA1) ...Distinct morphologies of the mitochondrial network support divergent metabolic and regulatory processes that determine cell function and fate. The mechanochemical GTPase optic atrophy 1 (OPA1) influences the architecture of cristae and catalyses the fusion of the mitochondrial inner membrane. Despite its fundamental importance, the molecular mechanisms by which OPA1 modulates mitochondrial morphology are unclear. Here, using a combination of cellular and structural analyses, we illuminate the molecular mechanisms that are key to OPA1-dependent membrane remodelling and fusion. Human OPA1 embeds itself into cardiolipin-containing membranes through a lipid-binding paddle domain. A conserved loop within the paddle domain inserts deeply into the bilayer, further stabilizing the interactions with cardiolipin-enriched membranes. OPA1 dimerization through the paddle domain promotes the helical assembly of a flexible OPA1 lattice on the membrane, which drives mitochondrial fusion in cells. Moreover, the membrane-bending OPA1 oligomer undergoes conformational changes that pull the membrane-inserting loop out of the outer leaflet and contribute to the mechanics of membrane remodelling. Our findings provide a structural framework for understanding how human OPA1 shapes mitochondrial morphology and show us how human disease mutations compromise OPA1 functions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26984.map.gz emd_26984.map.gz | 224.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26984-v30.xml emd-26984-v30.xml emd-26984.xml emd-26984.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26984_fsc.xml emd_26984_fsc.xml | 15 KB | Display |  FSC data file FSC data file |
| Images |  emd_26984.png emd_26984.png | 158.6 KB | ||
| Masks |  emd_26984_msk_1.map emd_26984_msk_1.map | 282.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26984.cif.gz emd-26984.cif.gz | 6.2 KB | ||
| Others |  emd_26984_additional_1.map.gz emd_26984_additional_1.map.gz emd_26984_half_map_1.map.gz emd_26984_half_map_1.map.gz emd_26984_half_map_2.map.gz emd_26984_half_map_2.map.gz | 259.5 MB 225.1 MB 225.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26984 http://ftp.pdbj.org/pub/emdb/structures/EMD-26984 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26984 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26984 | HTTPS FTP |
-Validation report
| Summary document |  emd_26984_validation.pdf.gz emd_26984_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26984_full_validation.pdf.gz emd_26984_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_26984_validation.xml.gz emd_26984_validation.xml.gz | 22 KB | Display | |
| Data in CIF |  emd_26984_validation.cif.gz emd_26984_validation.cif.gz | 28.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26984 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26984 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26984 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26984 | HTTPS FTP |
-Related structure data
| Related structure data |  8ct9MC  8ct1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26984.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26984.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refine3D map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.666 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26984_msk_1.map emd_26984_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Postprocess map
| File | emd_26984_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocess map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Refine3D half map 1
| File | emd_26984_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refine3D half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Refine3D half map 2
| File | emd_26984_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refine3D half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : OPA1
| Entire | Name: OPA1 |
|---|---|
| Components |
|
-Supramolecule #1: OPA1
| Supramolecule | Name: OPA1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Uniprot ID: O60313 - HUMAN OPA1, Dynamin-like 120 kDa protein, mitochondrial |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 820 KDa |
-Macromolecule #1: Dynamin-like 120 kDa protein, mitochondrial
| Macromolecule | Name: Dynamin-like 120 kDa protein, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 34 / Enantiomer: LEVO / EC number: dynamin GTPase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 111.804789 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MWRLRRAAVA CEVCQSLVKH SSGIKGSLPL QKLHLVSRSI YHSHHPTLKL QRPQLRTSFQ QFSSLTNLPL RKLKFSPIKY GYQPRRNFW PARLATRLLK LRYLILGSAV GGGYTAKKTF DQWKDMIPDL SEYKWIVPDI VWEIDEYIDF EKIRKALPSS E DLVKLAPD ...String: MWRLRRAAVA CEVCQSLVKH SSGIKGSLPL QKLHLVSRSI YHSHHPTLKL QRPQLRTSFQ QFSSLTNLPL RKLKFSPIKY GYQPRRNFW PARLATRLLK LRYLILGSAV GGGYTAKKTF DQWKDMIPDL SEYKWIVPDI VWEIDEYIDF EKIRKALPSS E DLVKLAPD FDKIVESLSL LKDFFTSGSP EETAFRATDR GSESDKHFRK VSDKEKIDQL QEELLHTQLK YQRILERLEK EN KELRKLV LQKDDKGIHH RKLKKSLIDM YSEVLDVLSD YDASYNTQDH LPRVVVVGDQ SAGKTSVLEM IAQARIFPRG SGE MMTRSP VKVTLSEGPH HVALFKDSSR EFDLTKEEDL AALRHEIELR MRKNVKEGCT VSPETISLNV KGPGLQRMVL VDLP GVINT VTSGMAPDTK ETIFSISKAY MQNPNAIILC IQDGSVDAER SIVTDLVSQM DPHGRRTIFV LTKVDLAEKN VASPS RIQQ IIEGKLFPMK ALGYFAVVTG KGNSSESIEA IREYEEEFFQ NSKLLKTSML KAHQVTTRNL SLAVSDCFWK MVRESV EQQ ADSFKATRFN LETEWKNNYP RLRELDRNEL FEKAKNEILD EVISLSQVTP KHWEEILQQS LWERVSTHVI ENIYLPA AQ TMNSGTFNTT VDIKLKQWTD KQLPNKAVEV AWETLQEEFS RFMTEPKGKE HDDIFDKLKE AVKEESIKRH KWNDFAED S LRVIQHNALE DRSISDKQQW DAAIYFMEEA LQARLKDTEN AIENMVGPDW KKRWLYWKNR TQEQCVHNET KNELEKMLK CNEEHPAYLA SDEITTVRKN LESRGVEVDP SLIKDTWHQV YRRHFLKTAL NHCNLCRRGF YYYQRHFVDS ELECNDVVLF WRIQRMLAI TANTLRQQLT NTEVRRLEKN VKEVLEDFAE DGEKKIKLLT GKRVQLAEDL KKVREIQEKL DAFIEALHQE K UniProtKB: Dynamin-like GTPase OPA1, mitochondrial |
-Macromolecule #2: CARDIOLIPIN
| Macromolecule | Name: CARDIOLIPIN / type: ligand / ID: 2 / Number of copies: 91 / Formula: CDL |
|---|---|
| Molecular weight | Theoretical: 1.464043 KDa |
| Chemical component information |  ChemComp-CDL: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 2.0 sec. / Average electron dose: 82.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)