[English] 日本語
 Yorodumi
Yorodumi- EMDB-26603: Structure of vesicular stomatitis virus (local reconstruction, 3.... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of vesicular stomatitis virus (local reconstruction, 3.5 A resolution) | |||||||||
 Map data Map data | Final sharpened | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Vesicular stomatitis virus / bullet-shaped particle / Rhrabdovirus / nucleocapsid / helical reconstruction / cryo electron microscopy / nucleoprotein / matrix protein / RNA / assembly / VIRAL PROTEIN-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host transcription initiation from RNA polymerase II promoter / RNA replication / host cell nuclear membrane / helical viral capsid / viral budding via host ESCRT complex / viral transcription / symbiont-mediated suppression of host mRNA export from nucleus / viral nucleocapsid / host cell cytoplasm / structural constituent of virion ...symbiont-mediated suppression of host transcription initiation from RNA polymerase II promoter / RNA replication / host cell nuclear membrane / helical viral capsid / viral budding via host ESCRT complex / viral transcription / symbiont-mediated suppression of host mRNA export from nucleus / viral nucleocapsid / host cell cytoplasm / structural constituent of virion / ribonucleoprotein complex / phosphorylation / viral envelope / virion membrane / RNA binding / membrane Similarity search - Function | |||||||||
| Biological species |  Vesicular stomatitis Indiana virus Vesicular stomatitis Indiana virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Jenni S / Horwitz JA / Bloyet L-M / Whelan SPJ / Harrison SC | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Visualizing molecular interactions that determine assembly of a bullet-shaped vesicular stomatitis virus particle. Authors: Simon Jenni / Joshua A Horwitz / Louis-Marie Bloyet / Sean P J Whelan / Stephen C Harrison /  Abstract: Vesicular stomatitis virus (VSV) is a negative-strand RNA virus with a non-segmented genome, closely related to rabies virus. Both have characteristic bullet-like shapes. We report the structure of ...Vesicular stomatitis virus (VSV) is a negative-strand RNA virus with a non-segmented genome, closely related to rabies virus. Both have characteristic bullet-like shapes. We report the structure of intact, infectious VSV particles determined by cryogenic electron microscopy. By compensating for polymorphism among viral particles with computational classification, we obtained a reconstruction of the shaft ("trunk") at 3.5 Å resolution, with lower resolution for the rounded tip. The ribonucleoprotein (RNP), genomic RNA complexed with nucleoprotein (N), curls into a dome-like structure with about eight gradually expanding turns before transitioning into the regular helical trunk. Two layers of matrix (M) protein link the RNP with the membrane. Radial inter-layer subunit contacts are fixed within single RNA-N-M1-M2 modules, but flexible lateral and axial interactions allow assembly of polymorphic virions. Together with published structures of recombinant N in various states, our results suggest a mechanism for membrane-coupled self-assembly of VSV and its relatives. #1:  Journal: Biorxiv / Year: 2022 Journal: Biorxiv / Year: 2022Title: Visualizing Molecular Interactions that Determine Assembly of a Bullet-Shaped Vesicular Stomatitis Virus Particle Authors: Jenni S / Horwitz JA / Bloyet L-M / Whelan SPJ / Harrison SC | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26603.map.gz emd_26603.map.gz | 59.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26603-v30.xml emd-26603-v30.xml emd-26603.xml emd-26603.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26603_fsc.xml emd_26603_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_26603.png emd_26603.png | 80.2 KB | ||
| Masks |  emd_26603_msk_1.map emd_26603_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26603.cif.gz emd-26603.cif.gz | 6 KB | ||
| Others |  emd_26603_additional_1.map.gz emd_26603_additional_1.map.gz emd_26603_half_map_1.map.gz emd_26603_half_map_1.map.gz emd_26603_half_map_2.map.gz emd_26603_half_map_2.map.gz | 49.2 MB 49.3 MB 49.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26603 http://ftp.pdbj.org/pub/emdb/structures/EMD-26603 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26603 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26603 | HTTPS FTP |
-Validation report
| Summary document |  emd_26603_validation.pdf.gz emd_26603_validation.pdf.gz | 767 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26603_full_validation.pdf.gz emd_26603_full_validation.pdf.gz | 766.5 KB | Display | |
| Data in XML |  emd_26603_validation.xml.gz emd_26603_validation.xml.gz | 16.7 KB | Display | |
| Data in CIF |  emd_26603_validation.cif.gz emd_26603_validation.cif.gz | 21.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26603 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26603 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26603 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26603 | HTTPS FTP |
-Related structure data
| Related structure data |  7umlMC  7umkC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26603.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26603.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final sharpened | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26603_msk_1.map emd_26603_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
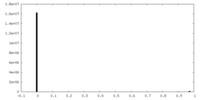
| Density Histograms |
-Additional map: Final
| File | emd_26603_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final | ||||||||||||
| Projections & Slices |
| ||||||||||||
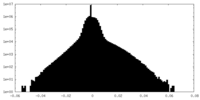
| Density Histograms |
-Half map: Half map 1
| File | emd_26603_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
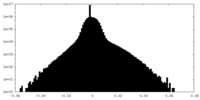
| Density Histograms |
-Half map: Half map 2
| File | emd_26603_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
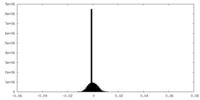
| Density Histograms |
- Sample components
Sample components
-Entire : Vesicular stomatitis Indiana virus
| Entire | Name:  Vesicular stomatitis Indiana virus Vesicular stomatitis Indiana virus |
|---|---|
| Components |
|
-Supramolecule #1: Vesicular stomatitis Indiana virus
| Supramolecule | Name: Vesicular stomatitis Indiana virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 11277 / Sci species name: Vesicular stomatitis Indiana virus / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|
-Macromolecule #1: Nucleoprotein
| Macromolecule | Name: Nucleoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vesicular stomatitis Indiana virus Vesicular stomatitis Indiana virus |
| Molecular weight | Theoretical: 47.463949 KDa |
| Recombinant expression | Organism:  Mesocricetus auratus (golden hamster) Mesocricetus auratus (golden hamster) |
| Sequence | String: MSVTVKRIID NTVIVPKLPA NEDPVEYPAD YFRKSKEIPL YINTTKSLSD LRGYVYQGLK SGNVSIIHVN SYLYGALKDI RGKLDKDWS SFGINIGKAG DTIGIFDLVS LKALDGVLPD GVSDASRTSA DDKWLPLYLL GLYRVGRTQM PEYRKKLMDG L TNQCKMIN ...String: MSVTVKRIID NTVIVPKLPA NEDPVEYPAD YFRKSKEIPL YINTTKSLSD LRGYVYQGLK SGNVSIIHVN SYLYGALKDI RGKLDKDWS SFGINIGKAG DTIGIFDLVS LKALDGVLPD GVSDASRTSA DDKWLPLYLL GLYRVGRTQM PEYRKKLMDG L TNQCKMIN EQFEPLVPEG RDIFDVWGND SNYTKIVAAV DMFFHMFKKH ECASFRYGTI VSRFKDCAAL ATFGHLCKIT GM STEDVTT WILNREVADE MVQMMLPGQE IDKADSYMPY LIDFGLSSKS PYSSVKNPAF HFWGQLTALL LRSTRARNAR QPD DIEYTS LTTAGLLYAY AVGSSADLAQ QFCVGDNKYT PDDSTGGLTT NAPPQGRDVV EWLGWFEDQN RKPTPDMMQY AKRA VMSLQ GLREKTIGKY AKSEFDK UniProtKB: Nucleoprotein |
-Macromolecule #2: Matrix protein
| Macromolecule | Name: Matrix protein / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vesicular stomatitis Indiana virus Vesicular stomatitis Indiana virus |
| Molecular weight | Theoretical: 26.103055 KDa |
| Recombinant expression | Organism:  Mesocricetus auratus (golden hamster) Mesocricetus auratus (golden hamster) |
| Sequence | String: MSSLKKILGL KGKGKKSKKL GIAPPPYEED TSMEYAPSAP IDKSYFGVDE MDTYDPNQLR YEKFFFTVKM TVRSNRPFRT YSDVAAAVS HWDHMYIGMA GKRPFYKILA FLGSSNLKAT PAVLADQGQP EYHAHCEGRA YLPHRMGKTP PMLNVPEHFR R PFNIGLYK ...String: MSSLKKILGL KGKGKKSKKL GIAPPPYEED TSMEYAPSAP IDKSYFGVDE MDTYDPNQLR YEKFFFTVKM TVRSNRPFRT YSDVAAAVS HWDHMYIGMA GKRPFYKILA FLGSSNLKAT PAVLADQGQP EYHAHCEGRA YLPHRMGKTP PMLNVPEHFR R PFNIGLYK GTIELTMTIY DDESLEAAPM IWDHFNSSKF SDFREKALMF GLIVEKKASG AWVLDSISHF K UniProtKB: Matrix protein |
-Macromolecule #3: RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3')
| Macromolecule | Name: RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3') type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Vesicular stomatitis Indiana virus Vesicular stomatitis Indiana virus |
| Molecular weight | Theoretical: 3.935198 KDa |
| Sequence | String: UUUUUUUUUU UUU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Details | Subparticle reconstruction |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-7uml: |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X