+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of human LACTB filament | |||||||||
 Map data Map data | Refine3D map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | serine protease / filament / tumor suppressor / membrane / mitochondria / beta lactamase / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationHydrolases; Acting on peptide bonds (peptidases) / regulation of lipid metabolic process / lipid metabolic process / peptidase activity / mitochondrion / proteolysis / identical protein binding / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
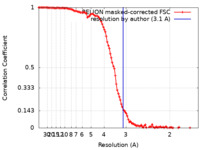
| Method | helical reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Bennett JA / Steward LR / Aydin H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Biol / Year: 2022 Journal: PLoS Biol / Year: 2022Title: The structure of the human LACTB filament reveals the mechanisms of assembly and membrane binding. Authors: Jeremy A Bennett / Lottie R Steward / Johannes Rudolph / Adam P Voss / Halil Aydin /  Abstract: Mitochondria are complex organelles that play a central role in metabolism. Dynamic membrane-associated processes regulate mitochondrial morphology and bioenergetics in response to cellular demand. ...Mitochondria are complex organelles that play a central role in metabolism. Dynamic membrane-associated processes regulate mitochondrial morphology and bioenergetics in response to cellular demand. In tumor cells, metabolic reprogramming requires active mitochondrial metabolism for providing key metabolites and building blocks for tumor growth and rapid proliferation. To counter this, the mitochondrial serine beta-lactamase-like protein (LACTB) alters mitochondrial lipid metabolism and potently inhibits the proliferation of a variety of tumor cells. Mammalian LACTB is localized in the mitochondrial intermembrane space (IMS), where it assembles into filaments to regulate the efficiency of essential metabolic processes. However, the structural basis of LACTB polymerization and regulation remains incompletely understood. Here, we describe how human LACTB self-assembles into micron-scale filaments that increase their catalytic activity. The electron cryo-microscopy (cryoEM) structure defines the mechanism of assembly and reveals how highly ordered filament bundles stabilize the active state of the enzyme. We identify and characterize residues that are located at the filament-forming interface and further show that mutations that disrupt filamentation reduce enzyme activity. Furthermore, our results provide evidence that LACTB filaments can bind lipid membranes. These data reveal the detailed molecular organization and polymerization-based regulation of human LACTB and provide new insights into the mechanism of mitochondrial membrane organization that modulates lipid metabolism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26595.map.gz emd_26595.map.gz | 98.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26595-v30.xml emd-26595-v30.xml emd-26595.xml emd-26595.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26595_fsc.xml emd_26595_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_26595.png emd_26595.png | 101.4 KB | ||
| Masks |  emd_26595_msk_1.map emd_26595_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26595.cif.gz emd-26595.cif.gz | 5.6 KB | ||
| Others |  emd_26595_additional_1.map.gz emd_26595_additional_1.map.gz emd_26595_half_map_1.map.gz emd_26595_half_map_1.map.gz emd_26595_half_map_2.map.gz emd_26595_half_map_2.map.gz | 116.6 MB 98.6 MB 98.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26595 http://ftp.pdbj.org/pub/emdb/structures/EMD-26595 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26595 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26595 | HTTPS FTP |
-Validation report
| Summary document |  emd_26595_validation.pdf.gz emd_26595_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26595_full_validation.pdf.gz emd_26595_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_26595_validation.xml.gz emd_26595_validation.xml.gz | 18.6 KB | Display | |
| Data in CIF |  emd_26595_validation.cif.gz emd_26595_validation.cif.gz | 24.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26595 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26595 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26595 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26595 | HTTPS FTP |
-Related structure data
| Related structure data |  7ulwMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26595.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26595.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refine3D map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8211 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26595_msk_1.map emd_26595_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Postprocess map
| File | emd_26595_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocess map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Refine3D half map 1
| File | emd_26595_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refine3D half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Refine3D half map 2
| File | emd_26595_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refine3D half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : LACTB
| Entire | Name: LACTB |
|---|---|
| Components |
|
-Supramolecule #1: LACTB
| Supramolecule | Name: LACTB / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Uniprot ID: P83111 - HUMAN LACTB, Serine beta-lactamase-like protein LACTB, mitochondrial |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 510 KDa |
-Macromolecule #1: Serine beta-lactamase-like protein LACTB, mitochondrial
| Macromolecule | Name: Serine beta-lactamase-like protein LACTB, mitochondrial type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: Hydrolases; Acting on peptide bonds (peptidases) |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 51.318648 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: APPCSRCFAR AIESSRDLLH RIKDEVGAPG IVVGVSVDGK EVWSEGLGYA DVENRVPCKP ETVMRIASIS KSLTMVALAK LWEAGKLDL DIPVQHYVPE FPEKEYEGEK VSVTTRLLIS HLSGIRHYEK DIKKVKEEKA YKALKMMKEN VAFEQEKEGK S NEKNDFTK ...String: APPCSRCFAR AIESSRDLLH RIKDEVGAPG IVVGVSVDGK EVWSEGLGYA DVENRVPCKP ETVMRIASIS KSLTMVALAK LWEAGKLDL DIPVQHYVPE FPEKEYEGEK VSVTTRLLIS HLSGIRHYEK DIKKVKEEKA YKALKMMKEN VAFEQEKEGK S NEKNDFTK FKTEQENEAK CRNSKPGKKK NDFEQGELYL REKFENSIES LRLFKNDPLF FKPGSQFLYS TFGYTLLAAI VE RASGCKY LDYMQKIFHD LDMLTTVQEE NEPVIYNRAR FYVYNKKKRL VNTPYVDNSY KWAGGGFLST VGDLLKFGNA MLY GYQVGL FKNSNENLLP GYLKPETMVM MWTPVPNTEM SWDKEGKYAM AWGVVERKQT YGSCRKQRHY ASHTGGAVGA SSVL LVLPE ELDTETINNK VPPRGIIVSI ICNMQSVGLN STALKIALEF DKDRSD UniProtKB: Serine beta-lactamase-like protein LACTB, mitochondrial |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 3.985 sec. / Average electron dose: 59.58 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)