+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | BamABCDE bound to substrate EspP | |||||||||
 Map data Map data | BAM-MBP-76EspP high resolution | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | membrane protein folding / membrane dynamics / outer membrane protein / BAM / beta-barrel / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationBam protein complex / Gram-negative-bacterium-type cell outer membrane assembly / protein insertion into membrane / Hydrolases; Acting on peptide bonds (peptidases); Serine endopeptidases / cell outer membrane / periplasmic space / serine-type endopeptidase activity / cell surface / proteolysis / extracellular region ...Bam protein complex / Gram-negative-bacterium-type cell outer membrane assembly / protein insertion into membrane / Hydrolases; Acting on peptide bonds (peptidases); Serine endopeptidases / cell outer membrane / periplasmic space / serine-type endopeptidase activity / cell surface / proteolysis / extracellular region / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
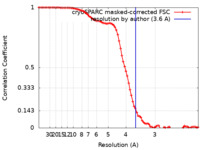
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Doyle MT / Jimah JR | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2022 Journal: Cell / Year: 2022Title: Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding. Authors: Matthew Thomas Doyle / John R Jimah / Tyrone Dowdy / Shannon I Ohlemacher / Mioara Larion / Jenny E Hinshaw / Harris D Bernstein /  Abstract: Transmembrane β barrel proteins are folded into the outer membrane (OM) of Gram-negative bacteria by the β barrel assembly machinery (BAM) via a poorly understood process that occurs without known ...Transmembrane β barrel proteins are folded into the outer membrane (OM) of Gram-negative bacteria by the β barrel assembly machinery (BAM) via a poorly understood process that occurs without known external energy sources. Here, we used single-particle cryo-EM to visualize the folding dynamics of a model β barrel protein (EspP) by BAM. We found that BAM binds the highly conserved "β signal" motif of EspP to correctly orient β strands in the OM during folding. We also found that the folding of EspP proceeds via "hybrid-barrel" intermediates in which membrane integrated β sheets are attached to the essential BAM subunit, BamA. The structures show an unprecedented deflection of the membrane surrounding the EspP intermediates and suggest that β sheets progressively fold toward BamA to form a β barrel. Along with in vivo experiments that tracked β barrel folding while the OM tension was modified, our results support a model in which BAM harnesses OM elasticity to accelerate β barrel folding. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26114.map.gz emd_26114.map.gz | 3.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26114-v30.xml emd-26114-v30.xml emd-26114.xml emd-26114.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26114_fsc.xml emd_26114_fsc.xml | 9 KB | Display |  FSC data file FSC data file |
| Images |  emd_26114.png emd_26114.png | 66.1 KB | ||
| Filedesc metadata |  emd-26114.cif.gz emd-26114.cif.gz | 12.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26114 http://ftp.pdbj.org/pub/emdb/structures/EMD-26114 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26114 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26114 | HTTPS FTP |
-Validation report
| Summary document |  emd_26114_validation.pdf.gz emd_26114_validation.pdf.gz | 375.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26114_full_validation.pdf.gz emd_26114_full_validation.pdf.gz | 375.1 KB | Display | |
| Data in XML |  emd_26114_validation.xml.gz emd_26114_validation.xml.gz | 10.6 KB | Display | |
| Data in CIF |  emd_26114_validation.cif.gz emd_26114_validation.cif.gz | 13.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26114 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26114 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26114 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26114 | HTTPS FTP |
-Related structure data
| Related structure data |  7ttcMC  7tszC  7tt0C  7tt1C  7tt2C  7tt3C  7tt4C  7tt5C  7tt6C  7tt7C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26114.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26114.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BAM-MBP-76EspP high resolution | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07382 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : BamABCDE bound to substrate EspP
| Entire | Name: BamABCDE bound to substrate EspP |
|---|---|
| Components |
|
-Supramolecule #1: BamABCDE bound to substrate EspP
| Supramolecule | Name: BamABCDE bound to substrate EspP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Outer membrane protein assembly factor BamB
| Macromolecule | Name: Outer membrane protein assembly factor BamB / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.882375 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: CSLFNSEEDV VKMSPLPTVE NQFTPTTAWS TSVGSGIGNF YSNLHPALAD NVVYAADRAG LVKALNADDG KEIWSVSLAE KDGWFSKEP ALLSGGVTVS GGHVYIGSEK AQVYALNTSD GTVAWQTKVA GEALSRPVVS DGLVLIHTSN GQLQALNEAD G AVKWTVNL ...String: CSLFNSEEDV VKMSPLPTVE NQFTPTTAWS TSVGSGIGNF YSNLHPALAD NVVYAADRAG LVKALNADDG KEIWSVSLAE KDGWFSKEP ALLSGGVTVS GGHVYIGSEK AQVYALNTSD GTVAWQTKVA GEALSRPVVS DGLVLIHTSN GQLQALNEAD G AVKWTVNL DMPSLSLRGE SAPTTAFGAA VVGGDNGRVS AVLMEQGQMI WQQRISQATG STEIDRLSDV DTTPVVVNGV VF ALAYNGN LTALDLRSGQ IMWKRELGSV NDFIVDGNRI YLVDQNDRVM ALTIDGGVTL WTQSDLLHRL LTSPVLYNGN LVV GDSEGY LHWINVEDGR FVAQQKVDSS GFQTEPVAAD GKLLIQAKDG TVYSITR UniProtKB: Outer membrane protein assembly factor BamB |
-Macromolecule #2: Outer membrane protein assembly factor BamA
| Macromolecule | Name: Outer membrane protein assembly factor BamA / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 89.750094 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AHHHHHHHHG GEGFVVKDIH FEGLQRVAVG AALLSMPVRT GDTVNDEDIS NTIRALFATG NFEDVRVLRD GDTLLVQVKE RPTIASITF SGNKSVKDDM LKQNLEASGV RVGESLDRTT IADIEKGLED FYYSVGKYSA SVKAVVTPLP RNRVDLKLVF Q EGVSAEIQ ...String: AHHHHHHHHG GEGFVVKDIH FEGLQRVAVG AALLSMPVRT GDTVNDEDIS NTIRALFATG NFEDVRVLRD GDTLLVQVKE RPTIASITF SGNKSVKDDM LKQNLEASGV RVGESLDRTT IADIEKGLED FYYSVGKYSA SVKAVVTPLP RNRVDLKLVF Q EGVSAEIQ QINIVGNHAF TTDELISHFQ LRDEVPWWNV VGDRKYQKQK LAGDLETLRS YYLDRGYARF NIDSTQVSLT PD KKGIYVT VNITEGDQYK LSGVEVSGNL AGHSAEIEQL TKIEPGELYN GTKVTKMEDD IKKLLGRYGY AYPRVQSMPE IND ADKTVK LRVNVDAGNR FYVRKIRFEG NDTSKDAVLR REMRQMEGAW LGSDLVDQGK ERLNRLGFFE TVDTDTQRVP GSPD QVDVV YKVKERNTGC FNFGIGYGTE SGVSFQAGVQ QDNWLGTGYA VGINGTKNDY QTYAELSVTN PYFTVDGVSL GGRLF YNDF QADDADLSDY TNKSYGTDVT LGFPINEYNS LRAGLGYVHN SLSNMQPQVA MWRYLYSMGE HPSTSDQDNS FKTDDF TFN YGWTYNKLDR GYFPTDGSRV NLTGKVTIPG SDNEYYKVTL DTATYVPIDD DHKWVVLGRT RWGYGDGLGG KEMPFYE NF YAGGSSTVRG FQSNTIGPKA VYFPHQASNY DPDYDYECAT QDGAKDLCKS DDAVGGNAMA VASLEFITPT PFISDKYA N SVRTSFFWDM GTVWDTNWDS SQYSGYPDYS DPSNIRMSAG IALQWMSPLG PLVFSYAQPF KKYDGDKAEQ FQFNIGKTW UniProtKB: Outer membrane protein assembly factor BamA |
-Macromolecule #3: Outer membrane protein assembly factor BamE
| Macromolecule | Name: Outer membrane protein assembly factor BamE / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.391554 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: CSTLERVVYR PDINQGNYLT ANDVSKIRVG MTQQQVAYAL GTPLMSDPFG TNTWFYVFRQ QPGHEGVTQQ TLTLTFNSSG VLTNIDNKP ALSGN UniProtKB: Outer membrane protein assembly factor BamE |
-Macromolecule #4: Outer membrane protein assembly factor BamD
| Macromolecule | Name: Outer membrane protein assembly factor BamD / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.816818 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: CSGSKEEVPD NPPNEIYATA QQKLQDGNWR QAITQLEALD NRYPFGPYSQ QVQLDLIYAY YKNADLPLAQ AAIDRFIRLN PTHPNIDYV MYMRGLTNMA LDDSALQGFF GVDRSDRDPQ HARAAFSDFS KLVRGYPNSQ YTTDATKRLV FLKDRLAKYE Y SVAEYYTE ...String: CSGSKEEVPD NPPNEIYATA QQKLQDGNWR QAITQLEALD NRYPFGPYSQ QVQLDLIYAY YKNADLPLAQ AAIDRFIRLN PTHPNIDYV MYMRGLTNMA LDDSALQGFF GVDRSDRDPQ HARAAFSDFS KLVRGYPNSQ YTTDATKRLV FLKDRLAKYE Y SVAEYYTE RGAWVAVVNR VEGMLRDYPD TQATRDALPL MENAYRQMQM NAQAEKVAKI IAANSSNT UniProtKB: Outer membrane protein assembly factor BamD |
-Macromolecule #5: Outer membrane protein assembly factor BamC
| Macromolecule | Name: Outer membrane protein assembly factor BamC / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.40125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: CSSDSRYKRQ VSGDEAYLEA APLAELHAPA GMILPVTSGD YAIPVTNGSG AVGKALDIRP PAQPLALVSG ARTQFTGDTA SLLVENGRG NTLWPQVVSV LQAKNYTITQ RDDAGQTLTT DWVQWNRLDE DEQYRGRYQI SVKPQGYQQA VTVKLLNLEQ A GKPVADAA ...String: CSSDSRYKRQ VSGDEAYLEA APLAELHAPA GMILPVTSGD YAIPVTNGSG AVGKALDIRP PAQPLALVSG ARTQFTGDTA SLLVENGRG NTLWPQVVSV LQAKNYTITQ RDDAGQTLTT DWVQWNRLDE DEQYRGRYQI SVKPQGYQQA VTVKLLNLEQ A GKPVADAA SMQRYSTEMM NVISAGLDKS ATDAANAAQN RASTTMDVQS AADDTGLPML VVRGPFNVVW QRLPAALEKV GM KVTDSTR SQGNMAVTYK PLSDSDWQEL GASDPGLASG DYKLQVGDLD NRSSLQFIDP KGHTLTQSQN DALVAVFQAA FSK UniProtKB: Outer membrane protein assembly factor BamC |
-Macromolecule #6: Serine protease EspP
| Macromolecule | Name: Serine protease EspP / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.379613 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: NLNKRMGDLR DINGEAGAWA RIMSGTGSAS GGFSDNYTHV QVGVDKKHEL DGLDLFTGFT VTHTDSSASA DVFSGKTKSV GAGLYASAM FDSGAYIDLI GKYVHHDNEY TATFAGLGTR DYSTHSWYAG AEAGYRYHVT EDAWIEPQAE LVYGSVSGKQ F AWKDQGMH ...String: NLNKRMGDLR DINGEAGAWA RIMSGTGSAS GGFSDNYTHV QVGVDKKHEL DGLDLFTGFT VTHTDSSASA DVFSGKTKSV GAGLYASAM FDSGAYIDLI GKYVHHDNEY TATFAGLGTR DYSTHSWYAG AEAGYRYHVT EDAWIEPQAE LVYGSVSGKQ F AWKDQGMH LSMKDKDYNP LIGRTGVDVG KSFSGKDWKV TARAGLGYQF DLLANGETVL RDASGEKRIK GEKDSRMLMS VG LNAEIRD NVRFGLEFEK SAFGKYNVDN AVNANFRYCF UniProtKB: Serine protease EspP |
-Macromolecule #7: 2,3-dideoxy-6-O-[2-deoxy-4-O-phosphono-2-(tetradecanoylamino)-alp...
| Macromolecule | Name: 2,3-dideoxy-6-O-[2-deoxy-4-O-phosphono-2-(tetradecanoylamino)-alpha-L-gulopyranosyl]-1-O-phosphono-beta-D-threo-hexopyranose type: ligand / ID: 7 / Number of copies: 1 / Formula: K33 |
|---|---|
| Molecular weight | Theoretical: 679.628 Da |
| Chemical component information | 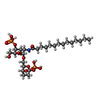 ChemComp-K33: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #0 - Average electron dose: 60.0 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K3 (6k x 4k) / #1 - Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.6 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)