[English] 日本語
 Yorodumi
Yorodumi- EMDB-25500: Kinetically trapped Pseudomonas-phage PaP3 portal protein - Full ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Kinetically trapped Pseudomonas-phage PaP3 portal protein - Full Length | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | portal protein / dodecamer / VIRAL PROTEIN | |||||||||
| Function / homology | ORF.04 Function and homology information Function and homology information | |||||||||
| Biological species |  Pseudomonas virus PaP3 Pseudomonas virus PaP3 | |||||||||
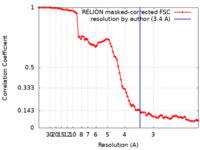
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Hou CFD / Swanson NA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2022 Journal: J Mol Biol / Year: 2022Title: Cryo-EM Structure of a Kinetically Trapped Dodecameric Portal Protein from the Pseudomonas-phage PaP3. Authors: Chun-Feng David Hou / Nicholas A Swanson / Fenglin Li / Ruoyu Yang / Ravi K Lokareddy / Gino Cingolani /  Abstract: Portal proteins are dodecameric assemblies that occupy a unique 5-fold vertex of the icosahedral capsid of tailed bacteriophages and herpesviruses. The portal vertex interrupts the icosahedral ...Portal proteins are dodecameric assemblies that occupy a unique 5-fold vertex of the icosahedral capsid of tailed bacteriophages and herpesviruses. The portal vertex interrupts the icosahedral symmetry, and in vivo, its assembly and incorporation in procapsid are controlled by the scaffolding protein. Ectopically expressed portal oligomers are polymorphic in solution, and portal rings built by a different number of subunits have been documented in the literature. In this paper, we describe the cryo-EM structure of the portal protein from the Pseudomonas-phage PaP3, which we determined at 3.4 Å resolution. Structural analysis revealed a dodecamer with helical rather than rotational symmetry, which we hypothesize is kinetically trapped. The helical assembly was stabilized by local mispairing of portal subunits caused by the slippage of crown and barrel helices that move like a lever with respect to the portal body. Removing the C-terminal barrel promoted assembly of undecameric and dodecameric rings with quasi-rotational symmetry, suggesting that the barrel contributes to subunits mispairing. However, ΔC-portal rings were intrinsically asymmetric, with most particles having one open portal subunit interface. Together, these data expand the structural repertoire of viral portal proteins to Pseudomonas-phages and shed light on the unexpected plasticity of the portal protein quaternary structure. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25500.map.gz emd_25500.map.gz | 40.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25500-v30.xml emd-25500-v30.xml emd-25500.xml emd-25500.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25500_fsc.xml emd_25500_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_25500.png emd_25500.png | 90.7 KB | ||
| Masks |  emd_25500_msk_1.map emd_25500_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-25500.cif.gz emd-25500.cif.gz | 5.7 KB | ||
| Others |  emd_25500_half_map_1.map.gz emd_25500_half_map_1.map.gz emd_25500_half_map_2.map.gz emd_25500_half_map_2.map.gz | 40.9 MB 40.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25500 http://ftp.pdbj.org/pub/emdb/structures/EMD-25500 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25500 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25500 | HTTPS FTP |
-Validation report
| Summary document |  emd_25500_validation.pdf.gz emd_25500_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25500_full_validation.pdf.gz emd_25500_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_25500_validation.xml.gz emd_25500_validation.xml.gz | 14.3 KB | Display | |
| Data in CIF |  emd_25500_validation.cif.gz emd_25500_validation.cif.gz | 20.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25500 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25500 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25500 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25500 | HTTPS FTP |
-Related structure data
| Related structure data |  7sxkMC  7syaC  7sz4C  7sz6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25500.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25500.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_25500_msk_1.map emd_25500_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_25500_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_25500_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Asymmetric dodecamer complex of phage PaP3 portal
| Entire | Name: Asymmetric dodecamer complex of phage PaP3 portal |
|---|---|
| Components |
|
-Supramolecule #1: Asymmetric dodecamer complex of phage PaP3 portal
| Supramolecule | Name: Asymmetric dodecamer complex of phage PaP3 portal / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Pseudomonas virus PaP3 Pseudomonas virus PaP3 |
-Macromolecule #1: Portal protein
| Macromolecule | Name: Portal protein / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas virus PaP3 Pseudomonas virus PaP3 |
| Molecular weight | Theoretical: 80.981344 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAKRRKIKPM DDEQVLRHLD QLVNDALDFN SSELSKQRSE ALKYYFGEPF GNERPGKSGI VSRDVQETVD WIMPSLMKVF TSGGQVVKY EPDTAEDVEQ AEQETEYVNY LFMRKNEGFK VMFDWFQDTL MMKTGVVKVY VEEVLKPTFE RFSGLSEDMV A DILSDPDT ...String: MAKRRKIKPM DDEQVLRHLD QLVNDALDFN SSELSKQRSE ALKYYFGEPF GNERPGKSGI VSRDVQETVD WIMPSLMKVF TSGGQVVKY EPDTAEDVEQ AEQETEYVNY LFMRKNEGFK VMFDWFQDTL MMKTGVVKVY VEEVLKPTFE RFSGLSEDMV A DILSDPDT SILAQSVDDD GTYTIKIRKD KKKREIKVLC VKPENFLVDR LATCIDDARF LCHREKYTVS DLRLLGVPED VI EELPYDE YEFSDSQPER LVRDNFDMTG QLQYNSGDDA EANREVWASE CYTLLDVDGD GISELRRILY VGDYIISNEP WDC RPFADL NAYRIAHKFH GMSVYDKIRD IQEIRSVLMR NIMDNIYRTN QGRSVVLDGQ VNLEDLLTNE AAGIVRVKSM NSIT PLETP QLSGEVYGML DRLEADRGKR TGITDRTRGL DQNTLHSNQA AMSVNQLMTA AEQQIDLIAR MFAETGVKRL FQLLH DHAI KYQNQEEVFQ LRGKWVAVNP ANWRERSDLT VTVGIGNMNK DQQMLHLMRI WEMAQAVVGG GGLGVLVSEQ NLYNIL KEV TENAGYKDPD RFWTNPNSPE ALQAKAIREQ KEAQPKPEDI KAQADAQRAQ SDALAKQAEA QMKQVEAQIR LAEIELK KQ EAVLQQREMA LKEAELQLER DRFTWERARN EAEYHLEATQ ARAAYIGDGK VPETKKPTKA VRR UniProtKB: ORF.04 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)