+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of SARS-CoV-2 spike in complex with polyclonal Fab14 | |||||||||
 Map data Map data | Sharpened map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
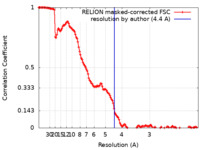
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Ward AB / Bangaru S / Antanasijevic A | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Structural mapping of antibody landscapes to human betacoronavirus spike proteins. Authors: Sandhya Bangaru / Aleksandar Antanasijevic / Nurgun Kose / Leigh M Sewall / Abigail M Jackson / Naveenchandra Suryadevara / Xiaoyan Zhan / Jonathan L Torres / Jeffrey Copps / Alba Torrents ...Authors: Sandhya Bangaru / Aleksandar Antanasijevic / Nurgun Kose / Leigh M Sewall / Abigail M Jackson / Naveenchandra Suryadevara / Xiaoyan Zhan / Jonathan L Torres / Jeffrey Copps / Alba Torrents de la Peña / James E Crowe / Andrew B Ward /  Abstract: Preexisting immunity against seasonal coronaviruses (CoVs) represents an important variable in predicting antibody responses and disease severity to severe acute respiratory syndrome CoV-2 (SARS-CoV- ...Preexisting immunity against seasonal coronaviruses (CoVs) represents an important variable in predicting antibody responses and disease severity to severe acute respiratory syndrome CoV-2 (SARS-CoV-2) infections. We used electron microscopy-based polyclonal epitope mapping (EMPEM) to characterize the antibody specificities against β-CoV spike proteins in prepandemic (PP) sera or SARS-CoV-2 convalescent (SC) sera. We observed that most PP sera had antibodies specific to seasonal human CoVs (HCoVs) OC43 and HKU1 spike proteins while the SC sera showed reactivity across all human β-CoVs. Detailed molecular mapping of spike-antibody complexes revealed epitopes that were differentially targeted by preexisting antibodies and SC serum antibodies. Our studies provide an antigenic landscape to β-HCoV spikes in the general population serving as a basis for cross-reactive epitope analyses in SARS-CoV-2-infected individuals. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24999.map.gz emd_24999.map.gz | 194.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24999-v30.xml emd-24999-v30.xml emd-24999.xml emd-24999.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24999_fsc.xml emd_24999_fsc.xml | 13.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_24999.png emd_24999.png | 90.3 KB | ||
| Masks |  emd_24999_msk_1.map emd_24999_msk_1.map | 209.3 MB |  Mask map Mask map | |
| Others |  emd_24999_half_map_1.map.gz emd_24999_half_map_1.map.gz emd_24999_half_map_2.map.gz emd_24999_half_map_2.map.gz | 166 MB 166.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24999 http://ftp.pdbj.org/pub/emdb/structures/EMD-24999 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24999 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24999 | HTTPS FTP |
-Validation report
| Summary document |  emd_24999_validation.pdf.gz emd_24999_validation.pdf.gz | 1003.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24999_full_validation.pdf.gz emd_24999_full_validation.pdf.gz | 1003 KB | Display | |
| Data in XML |  emd_24999_validation.xml.gz emd_24999_validation.xml.gz | 20.4 KB | Display | |
| Data in CIF |  emd_24999_validation.cif.gz emd_24999_validation.cif.gz | 26.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24999 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24999 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24999 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24999 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_24999.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24999.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_24999_msk_1.map emd_24999_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_24999_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_24999_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of SARS-CoV-2 spike in complex with polyclonal Fab14
| Entire | Name: Structure of SARS-CoV-2 spike in complex with polyclonal Fab14 |
|---|---|
| Components |
|
-Supramolecule #1: Structure of SARS-CoV-2 spike in complex with polyclonal Fab14
| Supramolecule | Name: Structure of SARS-CoV-2 spike in complex with polyclonal Fab14 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: 293F cells Homo sapiens (human) / Recombinant cell: 293F cells |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 / Component:
| ||||||
| Grid | Model: Quantifoil R1.2/1.3 / Pretreatment - Type: PLASMA CLEANING | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 5405 / Average exposure time: 7.5 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.7 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)