+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24676 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | AMC016 SOSIP.v4.2 in complex with PGV04 Fab | ||||||||||||
 Map data Map data | AMC016 SOSIP.v4.2 in complex with PGV04 Fab | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | HIV / Env / antibody / bnAb / VIRAL PROTEIN-IMMUNE SYSTEM complex | ||||||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
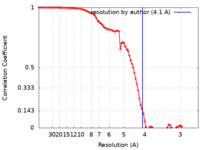
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | ||||||||||||
 Authors Authors | Bader DLV / Cottrell CA / Ward AB | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Virol / Year: 2022 Journal: J Virol / Year: 2022Title: The Glycan Hole Area of HIV-1 Envelope Trimers Contributes Prominently to the Induction of Autologous Neutralization. Authors: Anna Schorcht / Christopher A Cottrell / Pavel Pugach / Rajesh P Ringe / Alvin X Han / Joel D Allen / Tom L G M van den Kerkhof / Gemma E Seabright / Edith E Schermer / Thomas J Ketas / ...Authors: Anna Schorcht / Christopher A Cottrell / Pavel Pugach / Rajesh P Ringe / Alvin X Han / Joel D Allen / Tom L G M van den Kerkhof / Gemma E Seabright / Edith E Schermer / Thomas J Ketas / Judith A Burger / Jelle van Schooten / Celia C LaBranche / Gabriel Ozorowski / Natalia de Val / Daniel L V Bader / Hanneke Schuitemaker / Colin A Russell / David C Montefiori / Marit J van Gils / Max Crispin / P J Klasse / Andrew B Ward / John P Moore / Rogier W Sanders /    Abstract: The human immunodeficiency virus type 1 (HIV-1) trimeric envelope glycoprotein (Env) is heavily glycosylated, creating a dense glycan shield that protects the underlying peptidic surface from ...The human immunodeficiency virus type 1 (HIV-1) trimeric envelope glycoprotein (Env) is heavily glycosylated, creating a dense glycan shield that protects the underlying peptidic surface from antibody recognition. The absence of conserved glycans, due to missing potential N-linked glycosylation sites (PNGS), can result in strain-specific, autologous neutralizing antibody (NAb) responses. Here, we sought to gain a deeper understanding of the autologous neutralization by introducing holes in the otherwise dense glycan shields of the AMC011 and AMC016 SOSIP trimers. Specifically, when we knocked out the N130 and N289 glycans, which are absent from the well-characterized B41 SOSIP trimer, we observed stronger autologous NAb responses. We also analyzed the highly variable NAb responses induced in rabbits by diverse SOSIP trimers from subtypes A, B, and C. Statistical analysis, using linear regression, revealed that the cumulative area exposed on a trimer by glycan holes correlates with the magnitude of the autologous NAb response. Forty years after the first description of HIV-1, the search for a protective vaccine is still ongoing. The sole target for antibodies that can neutralize the virus are the trimeric envelope glycoproteins (Envs) located on the viral surface. The glycoprotein surface is covered with glycans that shield off the underlying protein components from recognition by the immune system. However, the Env trimers of some viral strains have holes in the glycan shield. Immunized animals developed antibodies against such glycan holes. These antibodies are generally strain specific. Here, we sought to gain a deeper understanding of what drives these specific immune responses. First, we show that strain-specific neutralizing antibody responses can be increased by creating artificial holes in the glycan shield. Second, when studying a diverse set of Env trimers with different characteristics, we found that the surface area of the glycan holes contributes prominently to the induction of strain-specific neutralizing antibodies. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24676.map.gz emd_24676.map.gz | 53.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24676-v30.xml emd-24676-v30.xml emd-24676.xml emd-24676.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24676_fsc.xml emd_24676_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_24676.png emd_24676.png | 62.1 KB | ||
| Filedesc metadata |  emd-24676.cif.gz emd-24676.cif.gz | 6.9 KB | ||
| Others |  emd_24676_half_map_1.map.gz emd_24676_half_map_1.map.gz emd_24676_half_map_2.map.gz emd_24676_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24676 http://ftp.pdbj.org/pub/emdb/structures/EMD-24676 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24676 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24676 | HTTPS FTP |
-Validation report
| Summary document |  emd_24676_validation.pdf.gz emd_24676_validation.pdf.gz | 815 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24676_full_validation.pdf.gz emd_24676_full_validation.pdf.gz | 814.5 KB | Display | |
| Data in XML |  emd_24676_validation.xml.gz emd_24676_validation.xml.gz | 16.2 KB | Display | |
| Data in CIF |  emd_24676_validation.cif.gz emd_24676_validation.cif.gz | 20.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24676 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24676 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24676 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24676 | HTTPS FTP |
-Related structure data
| Related structure data |  7rsoMC  7rsnC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_24676.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24676.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AMC016 SOSIP.v4.2 in complex with PGV04 Fab | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: AMC016 SOSIP.v4.2 in complex with PGV04 Fab
| File | emd_24676_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AMC016 SOSIP.v4.2 in complex with PGV04 Fab | ||||||||||||
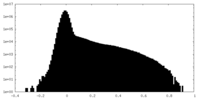
| Projections & Slices |
| ||||||||||||
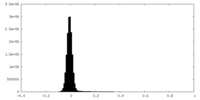
| Density Histograms |
-Half map: AMC016 SOSIP.v4.2 in complex with PGV04 Fab
| File | emd_24676_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AMC016 SOSIP.v4.2 in complex with PGV04 Fab | ||||||||||||
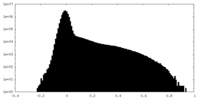
| Projections & Slices |
| ||||||||||||
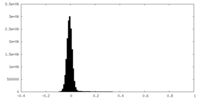
| Density Histograms |
- Sample components
Sample components
-Entire : AMC016 SOSIP.v4.2 in complex with PGV04 Fab
| Entire | Name: AMC016 SOSIP.v4.2 in complex with PGV04 Fab |
|---|---|
| Components |
|
-Supramolecule #1: AMC016 SOSIP.v4.2 in complex with PGV04 Fab
| Supramolecule | Name: AMC016 SOSIP.v4.2 in complex with PGV04 Fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: AMC016 gp120
| Macromolecule | Name: AMC016 gp120 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 54.890215 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AEEELWVTVY YGVPVWKEAT TTLFCASDAK AYDTEVRNVW ATHACVPTDP SPQEVVLANV TENFNMWKNN MVEQMHEDII SLWDQSLKP CVKLTPLCVT LNCTDLGNAT DAINRNTTDA PNSTLRTMEE KGEIKNCSFN ITTSVRDKMQ KEYATFYKLD I VPIDNDNN ...String: AEEELWVTVY YGVPVWKEAT TTLFCASDAK AYDTEVRNVW ATHACVPTDP SPQEVVLANV TENFNMWKNN MVEQMHEDII SLWDQSLKP CVKLTPLCVT LNCTDLGNAT DAINRNTTDA PNSTLRTMEE KGEIKNCSFN ITTSVRDKMQ KEYATFYKLD I VPIDNDNN SYRLINCNTS VITQACPKVS FEPIPIHYCA PAGFAILKCN NKTFNGTGPC TNVSTVQCTH GIRPVVSTQL LL NGSLAEE EIVIRSENFT DNAKTIIVQL NESVEINCTR PNNNTRKSIH IGPGRWFYTT GQIIGNIRQA HCNISRAKWN NTL HKIVKK LREQFRNKTI VFKQSSGGDP EIVMHSFNCG GEFFYCNSTQ LFNSTWYGNE SSDNPGVEGN ITLPCRIKQI INLW QEVGK AMYAPPIGGQ IRCSSNITGL LLTRDGGNNN ITTEIFRPGG GDMRDNWRSE LYKYKVVKIE PLGVAPTKCK RRVVQ RRRR RR |
-Macromolecule #2: AMC016 gp41
| Macromolecule | Name: AMC016 gp41 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.311746 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGTIGAMFL GFLGAAGSTM GAASMTLTVQ ARQLLSGIVQ QQSNLLRAPE AQQHLLKLTV WGIKQLQARV LAVERYLKDQ QLLGIWGCS GKLICCTAVP WNASWSNKSL DNIWNNMTWM EWEKEISNYT NLIYNLIEES QNQQEKNEQE LLELD |
-Macromolecule #3: PGV04 Fab heavy chain
| Macromolecule | Name: PGV04 Fab heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.759861 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGSG VKKPGASVRV SCWTSEDIFE RTELIHWVRQ APGQGLEWIG WVKTVTGAVN FGSPDFRQRV SLTRDRDLFT AHMDIRGLT QGDTATYFCA RQKFYTGGQG WYFDLWGRGT LIVVSSASTK GPSVFPLAPS SKSTSGGTAA LGCLVKDYFP E PVTVSWNS ...String: QVQLVQSGSG VKKPGASVRV SCWTSEDIFE RTELIHWVRQ APGQGLEWIG WVKTVTGAVN FGSPDFRQRV SLTRDRDLFT AHMDIRGLT QGDTATYFCA RQKFYTGGQG WYFDLWGRGT LIVVSSASTK GPSVFPLAPS SKSTSGGTAA LGCLVKDYFP E PVTVSWNS GALTSGVHTF PAVLQSSGLY SLSSVVTVPS SSLGTQTYIC NVNHKPSNTK VDKKVEPKSC D |
-Macromolecule #4: PGV04 kappa chain
| Macromolecule | Name: PGV04 kappa chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.073822 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPGT LSLSPGETAS LSCTAASYGH MTWYQKKPGQ PPKLLIFATS KRASGIPDRF SGSQFGKQYT LTITRMEPED FARYYCQQL EFFGQGTRLE IRRTVAAPSV FIFPPSDEQL KSGTASVVCL LNNFYPREAK VQWKVDNALQ SGNSQESVTE Q DSKDSTYS ...String: EIVLTQSPGT LSLSPGETAS LSCTAASYGH MTWYQKKPGQ PPKLLIFATS KRASGIPDRF SGSQFGKQYT LTITRMEPED FARYYCQQL EFFGQGTRLE IRRTVAAPSV FIFPPSDEQL KSGTASVVCL LNNFYPREAK VQWKVDNALQ SGNSQESVTE Q DSKDSTYS LSSTLTLSKA DYEKHKVYAC EVTHQGLSSP VTKSFNRGEC |
-Macromolecule #9: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 9 / Number of copies: 45 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: C-flat-2/2 |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 3801 / Average exposure time: 7.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7rso: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)