[English] 日本語
 Yorodumi
Yorodumi- EMDB-7568: Glutaraldehyde-treated BG505 SOSIP.664 Env in complex with PGV04 Fab -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7568 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Glutaraldehyde-treated BG505 SOSIP.664 Env in complex with PGV04 Fab | |||||||||||||||
 Map data Map data | Glutaraldehyde-treated BG505 SOSIP.664 Env in complex with PGV04 Fab | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | glutaraldehyde / BG505 / SOSIP / PGV04 / HIV / HIV-1 / trimer / immmune complex / immunogen / Env / cross link / B cell activation / Th cell activation / VIRAL PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / membrane Similarity search - Function | |||||||||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
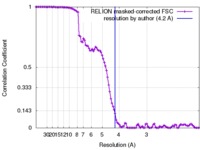
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||||||||
 Authors Authors | Pallesen J / de Val N / Ward AB | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2018 Journal: PLoS Pathog / Year: 2018Title: Structural and immunologic correlates of chemically stabilized HIV-1 envelope glycoproteins. Authors: Torben Schiffner / Jesper Pallesen / Rebecca A Russell / Jonathan Dodd / Natalia de Val / Celia C LaBranche / David Montefiori / Georgia D Tomaras / Xiaoying Shen / Scarlett L Harris / Amin ...Authors: Torben Schiffner / Jesper Pallesen / Rebecca A Russell / Jonathan Dodd / Natalia de Val / Celia C LaBranche / David Montefiori / Georgia D Tomaras / Xiaoying Shen / Scarlett L Harris / Amin E Moghaddam / Oleksandr Kalyuzhniy / Rogier W Sanders / Laura E McCoy / John P Moore / Andrew B Ward / Quentin J Sattentau /    Abstract: Inducing broad spectrum neutralizing antibodies against challenging pathogens such as HIV-1 is a major vaccine design goal, but may be hindered by conformational instability within viral envelope ...Inducing broad spectrum neutralizing antibodies against challenging pathogens such as HIV-1 is a major vaccine design goal, but may be hindered by conformational instability within viral envelope glycoproteins (Env). Chemical cross-linking is widely used for vaccine antigen stabilization, but how this process affects structure, antigenicity and immunogenicity is poorly understood and its use remains entirely empirical. We have solved the first cryo-EM structure of a cross-linked vaccine antigen. The 4.2 Å structure of HIV-1 BG505 SOSIP soluble recombinant Env in complex with a CD4 binding site-specific broadly neutralizing antibody (bNAb) Fab fragment reveals how cross-linking affects key properties of the trimer. We observed density corresponding to highly specific glutaraldehyde (GLA) cross-links between gp120 monomers at the trimer apex and between gp120 and gp41 at the trimer interface that had strikingly little impact on overall trimer conformation, but critically enhanced trimer stability and improved Env antigenicity. Cross-links were also observed within gp120 at sites associated with the N241/N289 glycan hole that locally modified trimer antigenicity. In immunogenicity studies, the neutralizing antibody response to cross-linked trimers showed modest but significantly greater breadth against a global panel of difficult-to-neutralize Tier-2 heterologous viruses. Moreover, the specificity of autologous Tier-2 neutralization was modified away from the N241/N289 glycan hole, implying a novel specificity. Finally, we have investigated for the first time T helper cell responses to next-generation soluble trimers, and report on vaccine-relevant immunodominant responses to epitopes within BG505 that are modified by cross-linking. Elucidation of the structural correlates of a cross-linked viral glycoprotein will allow more rational use of this methodology for vaccine design, and reveals a strategy with promise for eliciting neutralizing antibodies needed for an effective HIV-1 vaccine. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7568.map.gz emd_7568.map.gz | 96.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7568-v30.xml emd-7568-v30.xml emd-7568.xml emd-7568.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7568_fsc.xml emd_7568_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_7568.png emd_7568.png | 86.2 KB | ||
| Filedesc metadata |  emd-7568.cif.gz emd-7568.cif.gz | 6.9 KB | ||
| Others |  emd_7568_half_map_1.map.gz emd_7568_half_map_1.map.gz emd_7568_half_map_2.map.gz emd_7568_half_map_2.map.gz | 80.6 MB 80.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7568 http://ftp.pdbj.org/pub/emdb/structures/EMD-7568 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7568 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7568 | HTTPS FTP |
-Related structure data
| Related structure data |  6crqMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7568.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7568.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Glutaraldehyde-treated BG505 SOSIP.664 Env in complex with PGV04 Fab | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.02 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Glutaraldehyde-treated BG505 SOSIP.664 Env in complex with PGV04 Fab
| File | emd_7568_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Glutaraldehyde-treated BG505 SOSIP.664 Env in complex with PGV04 Fab | ||||||||||||
| Projections & Slices |
| ||||||||||||
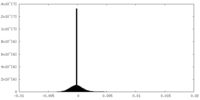
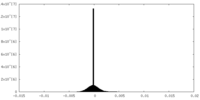
| Density Histograms |
-Half map: Glutaraldehyde-treated BG505 SOSIP.664 Env in complex with PGV04 Fab
| File | emd_7568_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Glutaraldehyde-treated BG505 SOSIP.664 Env in complex with PGV04 Fab | ||||||||||||
| Projections & Slices |
| ||||||||||||
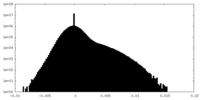
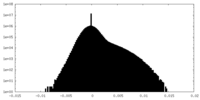
| Density Histograms |
- Sample components
Sample components
-Entire : Glutaraldehyde-treated BG505 SOSIP.664 Env in complex with PGV04 Fab
| Entire | Name: Glutaraldehyde-treated BG505 SOSIP.664 Env in complex with PGV04 Fab |
|---|---|
| Components |
|
-Supramolecule #1: Glutaraldehyde-treated BG505 SOSIP.664 Env in complex with PGV04 Fab
| Supramolecule | Name: Glutaraldehyde-treated BG505 SOSIP.664 Env in complex with PGV04 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 550 KDa |
-Macromolecule #1: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 53.950172 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ENLWVTVYYG VPVWKDAETT LFCASDAKAY ETEKHNVWAT HACVPTDPNP QEIHLENVTE EFNMWKNNMV EQMHTDIISL WDQSLKPCV KLTPLCVTLQ CTNVTNAITD DMRGELKNCS FNMTTELRDK KQKVYSLFYR LDVVQINENQ GNRSNNSNKE Y RLINCNTS ...String: ENLWVTVYYG VPVWKDAETT LFCASDAKAY ETEKHNVWAT HACVPTDPNP QEIHLENVTE EFNMWKNNMV EQMHTDIISL WDQSLKPCV KLTPLCVTLQ CTNVTNAITD DMRGELKNCS FNMTTELRDK KQKVYSLFYR LDVVQINENQ GNRSNNSNKE Y RLINCNTS AITQACPKVS FEPIPIHYCA PAGFAILKCK DKKFNGTGPC PSVSTVQCTH GIKPVVSTQL LLNGSLAEEE VM IRSENIT NNAKNILVQF NTPVQINCTR PNNNTRKSIR IGPGQAFYAT GDIIGDIRQA HCNVSKATWN ETLGKVVKQL RKH FGNNTI IRFANSSGGD LEVTTHSFNC GGEFFYCNTS GLFNSTWISN TSVQGSNSTG SNDSITLPCR IKQIINMWQR IGQA MYAPP IQGVIRCVSN ITGLILTRDG GSTNSTTETF RPGGGDMRDN WRSELYKYKV VKIEPLGVAP TRCKRRVVGR RRRRR UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.146482 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEA QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALD UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: PGV04 VH
| Macromolecule | Name: PGV04 VH / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.644771 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGSG VKKPGASVRV SCWTSEDIFE RTELIHWVRQ APGQGLEWIG WVKTVTGAVN FGSPDFRQRV SLTRDRDLFT AHMDIRGLT QGDTATYFCA RQKFYTGGQG WYFDLWGRGT LIVVSSASTK GPSVFPLAPS SKSTSGGTAA LGCLVKDYFP E PVTVSWNS ...String: QVQLVQSGSG VKKPGASVRV SCWTSEDIFE RTELIHWVRQ APGQGLEWIG WVKTVTGAVN FGSPDFRQRV SLTRDRDLFT AHMDIRGLT QGDTATYFCA RQKFYTGGQG WYFDLWGRGT LIVVSSASTK GPSVFPLAPS SKSTSGGTAA LGCLVKDYFP E PVTVSWNS GALTSGVHTF PAVLQSSGLY SLSSVVTVPS SSLGTQTYIC NVNHKPSNTK VDKKVEPKSC |
-Macromolecule #4: PGV04 VL
| Macromolecule | Name: PGV04 VL / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.073822 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPGT LSLSPGETAS LSCTAASYGH MTWYQKKPGQ PPKLLIFATS KRASGIPDRF SGSQFGKQYT LTITRMEPED FARYYCQQL EFFGQGTRLE IRRTVAAPSV FIFPPSDEQL KSGTASVVCL LNNFYPREAK VQWKVDNALQ SGNSQESVTE Q DSKDSTYS ...String: EIVLTQSPGT LSLSPGETAS LSCTAASYGH MTWYQKKPGQ PPKLLIFATS KRASGIPDRF SGSQFGKQYT LTITRMEPED FARYYCQQL EFFGQGTRLE IRRTVAAPSV FIFPPSDEQL KSGTASVVCL LNNFYPREAK VQWKVDNALQ SGNSQESVTE Q DSKDSTYS LSSTLTLSKA DYEKHKVYAC EVTHQGLSSP VTKSFNRGEC |
-Macromolecule #9: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 9 / Number of copies: 18 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.67 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Name: TBS / Details: 50 mM Tris, 150 mM NaCl, 0.3 mM DDM. |
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 297 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-35 / Number grids imaged: 1 / Number real images: 1329 / Average exposure time: 7.0 sec. / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-6crq: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)