[日本語] English
 万見
万見- EMDB-2462: Structure and Host Adhesion Mechanism of Virulent Lactococcal Phage p2 -
+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-2462 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
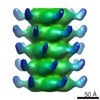
| タイトル | Structure and Host Adhesion Mechanism of Virulent Lactococcal Phage p2 | |||||||||
 マップデータ マップデータ | Icosahedral reconstruction of the capsid of lactococcal phage p2. | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | Lactococcus lactis / Siphoviridae / 936 phages / Bacteriophage / electron microscopy / single-particle / p2 | |||||||||
| 生物種 |  Lactococcus lactis phage p2 (ウイルス) Lactococcus lactis phage p2 (ウイルス) | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 13.0 Å | |||||||||
 データ登録者 データ登録者 | Bebeacua C / Tremblay D / Farenc C / Chapot MP / Sadovskaya I / van Heel M / Veesler D / Moineau S / Cambillau C | |||||||||
 引用 引用 |  ジャーナル: J Virol / 年: 2013 ジャーナル: J Virol / 年: 2013タイトル: Structure, adsorption to host, and infection mechanism of virulent lactococcal phage p2. 著者: Cecilia Bebeacua / Denise Tremblay / Carine Farenc / Marie-Pierre Chapot-Chartier / Irina Sadovskaya / Marin van Heel / David Veesler / Sylvain Moineau / Christian Cambillau /  要旨: Lactococcal siphophages from the 936 and P335 groups infect the Gram-positive bacterium Lactococcus lactis using receptor binding proteins (RBPs) attached to their baseplate, a large multiprotein ...Lactococcal siphophages from the 936 and P335 groups infect the Gram-positive bacterium Lactococcus lactis using receptor binding proteins (RBPs) attached to their baseplate, a large multiprotein complex at the distal part of the tail. We have previously reported the crystal and electron microscopy (EM) structures of the baseplates of phages p2 (936 group) and TP901-1 (P335 group) as well as the full EM structure of the TP901-1 virion. Here, we report the complete EM structure of siphophage p2, including its capsid, connector complex, tail, and baseplate. Furthermore, we show that the p2 tail is characterized by the presence of protruding decorations, which are related to adhesins and are likely contributed by the major tail protein C-terminal domains. This feature is reminiscent of the tail of Escherichia coli phage λ and Bacillus subtilis phage SPP1 and might point to a common mechanism for establishing initial interactions with their bacterial hosts. Comparative analyses showed that the architecture of the phage p2 baseplate differs largely from that of lactococcal phage TP901-1. We quantified the interaction of its RBP with the saccharidic receptor and determined that specificity is due to lower k(off) values of the RBP/saccharidic dissociation. Taken together, these results suggest that the infection of L. lactis strains by phage p2 is a multistep process that involves reversible attachment, followed by baseplate activation, specific attachment of the RBPs to the saccharidic receptor, and DNA ejection. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_2462.map.gz emd_2462.map.gz | 5.6 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-2462-v30.xml emd-2462-v30.xml emd-2462.xml emd-2462.xml | 10 KB 10 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  EMD-2462.png EMD-2462.png emd_2462.png emd_2462.png | 190.2 KB 190.2 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2462 http://ftp.pdbj.org/pub/emdb/structures/EMD-2462 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2462 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2462 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_2462_validation.pdf.gz emd_2462_validation.pdf.gz | 235.6 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_2462_full_validation.pdf.gz emd_2462_full_validation.pdf.gz | 234.7 KB | 表示 | |
| XML形式データ |  emd_2462_validation.xml.gz emd_2462_validation.xml.gz | 6.1 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2462 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2462 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2462 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2462 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_2462.map.gz / 形式: CCP4 / 大きさ: 29.8 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_2462.map.gz / 形式: CCP4 / 大きさ: 29.8 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Icosahedral reconstruction of the capsid of lactococcal phage p2. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 3.53 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : Capsid of Lactococcal phage p2
| 全体 | 名称: Capsid of Lactococcal phage p2 |
|---|---|
| 要素 |
|
-超分子 #1000: Capsid of Lactococcal phage p2
| 超分子 | 名称: Capsid of Lactococcal phage p2 / タイプ: sample / ID: 1000 / 集合状態: 60-mer / Number unique components: 1 |
|---|---|
| 分子量 | 実験値: 3 MDa / 理論値: 3 MDa |
-超分子 #1: Lactococcus lactis phage p2
| 超分子 | 名称: Lactococcus lactis phage p2 / タイプ: virus / ID: 1 / Name.synonym: Lactococcal phage p2 詳細: The capsid was selected from a sample containing entire Lactococcal phages p2. NCBI-ID: 100641 / 生物種: Lactococcus lactis phage p2 / ウイルスタイプ: OTHER / ウイルス・単離状態: SPECIES / ウイルス・エンベロープ: No / ウイルス・中空状態: No / Syn species name: Lactococcal phage p2 |
|---|---|
| 宿主 | 生物種: Lactococcal lactis / 株: NZ9000 / 別称: BACTERIA(EUBACTERIA) |
| ウイルス殻 | Shell ID: 1 / 名称: T7 / 直径: 660 Å / T番号(三角分割数): 7 |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.5 / 詳細: 50 mM Tris-HCl, 100 mM NaCl, 8 mM MgSO4 |
|---|---|
| グリッド | 詳細: Quantifoil grids |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / 装置: FEI VITROBOT MARK I / 手法: Blot for 2 seconds before plunging |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI/PHILIPS CM200T |
|---|---|
| アライメント法 | Legacy - 非点収差: Objective lens astigmatism was corrected at 100,000 times magnification. |
| 日付 | 2009年7月1日 |
| 撮影 | カテゴリ: CCD フィルム・検出器のモデル: GENERIC TVIPS (4k x 4k) デジタル化 - サンプリング間隔: 3.53 µm / 実像数: 200 / 平均電子線量: 10 e/Å2 |
| 電子線 | 加速電圧: 200 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: SPOT SCAN / 撮影モード: BRIGHT FIELD / Cs: 2.2 mm / 最大 デフォーカス(公称値): 0.002 µm / 最小 デフォーカス(公称値): 0.001 µm / 倍率(公称値): 50000 |
| 試料ステージ | 試料ホルダーモデル: GATAN LIQUID NITROGEN |
- 画像解析
画像解析
| 詳細 | Reconstruction imposing Icosahedral reconstruction |
|---|---|
| CTF補正 | 詳細: Images |
| 最終 再構成 | 想定した対称性 - 点群: I (正20面体型対称) / アルゴリズム: OTHER / 解像度のタイプ: BY AUTHOR / 解像度: 13.0 Å / 解像度の算出法: OTHER / ソフトウェア - 名称: IMAGIC / 使用した粒子像数: 3329 |
-原子モデル構築 1
| 初期モデル | PDB ID: |
|---|---|
| ソフトウェア | 名称:  Chimera Chimera |
| 詳細 | 60 copies of the hexamer of HK97 MCP were manually fitted and automatically refined with Chimera |
| 精密化 | 空間: REAL / プロトコル: RIGID BODY FIT |
 ムービー
ムービー コントローラー
コントローラー













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)