[English] 日本語
 Yorodumi
Yorodumi- EMDB-2459: Structure and Host Adhesion Mechanism of Virulent Lactococcal Phage p2 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2459 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure and Host Adhesion Mechanism of Virulent Lactococcal Phage p2 | |||||||||
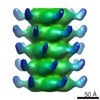
 Map data Map data | Icosahedral reconstruction of the capsid of p2 phage | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | EM / capsid / icosahedral / p2 / lactococcal phage | |||||||||
| Biological species |  Lactococcus phage p2 (virus) Lactococcus phage p2 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 13.0 Å | |||||||||
 Authors Authors | Bebeacua C / Tremblay D / Farenc C / Chapot MP / Sadovskaya I / van Heel M / Veesler D / Moineau S / Cambillau C | |||||||||
 Citation Citation |  Journal: J Virol / Year: 2013 Journal: J Virol / Year: 2013Title: Structure, adsorption to host, and infection mechanism of virulent lactococcal phage p2. Authors: Cecilia Bebeacua / Denise Tremblay / Carine Farenc / Marie-Pierre Chapot-Chartier / Irina Sadovskaya / Marin van Heel / David Veesler / Sylvain Moineau / Christian Cambillau /  Abstract: Lactococcal siphophages from the 936 and P335 groups infect the Gram-positive bacterium Lactococcus lactis using receptor binding proteins (RBPs) attached to their baseplate, a large multiprotein ...Lactococcal siphophages from the 936 and P335 groups infect the Gram-positive bacterium Lactococcus lactis using receptor binding proteins (RBPs) attached to their baseplate, a large multiprotein complex at the distal part of the tail. We have previously reported the crystal and electron microscopy (EM) structures of the baseplates of phages p2 (936 group) and TP901-1 (P335 group) as well as the full EM structure of the TP901-1 virion. Here, we report the complete EM structure of siphophage p2, including its capsid, connector complex, tail, and baseplate. Furthermore, we show that the p2 tail is characterized by the presence of protruding decorations, which are related to adhesins and are likely contributed by the major tail protein C-terminal domains. This feature is reminiscent of the tail of Escherichia coli phage λ and Bacillus subtilis phage SPP1 and might point to a common mechanism for establishing initial interactions with their bacterial hosts. Comparative analyses showed that the architecture of the phage p2 baseplate differs largely from that of lactococcal phage TP901-1. We quantified the interaction of its RBP with the saccharidic receptor and determined that specificity is due to lower k(off) values of the RBP/saccharidic dissociation. Taken together, these results suggest that the infection of L. lactis strains by phage p2 is a multistep process that involves reversible attachment, followed by baseplate activation, specific attachment of the RBPs to the saccharidic receptor, and DNA ejection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2459.map.gz emd_2459.map.gz | 23.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2459-v30.xml emd-2459-v30.xml emd-2459.xml emd-2459.xml | 10.3 KB 10.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2459.png emd_2459.png | 157.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2459 http://ftp.pdbj.org/pub/emdb/structures/EMD-2459 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2459 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2459 | HTTPS FTP |
-Validation report
| Summary document |  emd_2459_validation.pdf.gz emd_2459_validation.pdf.gz | 243.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2459_full_validation.pdf.gz emd_2459_full_validation.pdf.gz | 242.8 KB | Display | |
| Data in XML |  emd_2459_validation.xml.gz emd_2459_validation.xml.gz | 6.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2459 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2459 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2459 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2459 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2459.map.gz / Format: CCP4 / Size: 29.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2459.map.gz / Format: CCP4 / Size: 29.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Icosahedral reconstruction of the capsid of p2 phage | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Icosahedral capsid of the lactococcal phage p2
| Entire | Name: Icosahedral capsid of the lactococcal phage p2 |
|---|---|
| Components |
|
-Supramolecule #1000: Icosahedral capsid of the lactococcal phage p2
| Supramolecule | Name: Icosahedral capsid of the lactococcal phage p2 / type: sample / ID: 1000 Details: The sample corresponded to the full phage but only the capsid particles were selected. Oligomeric state: 60-mer / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 13 MDa / Theoretical: 13 MDa |
-Supramolecule #1: Lactococcus phage p2
| Supramolecule | Name: Lactococcus phage p2 / type: virus / ID: 1 Details: The sample contained the full phages with tail and baseplate. Only the capsids were selected. NCBI-ID: 254252 / Sci species name: Lactococcus phage p2 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Lactococcus lactis (lactic acid bacteria) / synonym: BACTERIA(EUBACTERIA) Lactococcus lactis (lactic acid bacteria) / synonym: BACTERIA(EUBACTERIA) |
| Molecular weight | Experimental: 13 MDa / Theoretical: 13 MDa |
| Virus shell | Shell ID: 1 / Diameter: 600 Å / T number (triangulation number): 7 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: SM buffer (50 mM Tris-HCl, 100 mM NaCl, 8 mM MgSO4) |
|---|---|
| Grid | Details: Quantifoil grids were glow discharged for 20 seconds |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 120 K / Instrument: FEI VITROBOT MARK I / Method: Blot for 2 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Temperature | Min: 80 K / Max: 105 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 130,000 times magnification. |
| Specialist optics | Energy filter - Name: FEI |
| Date | Feb 1, 2008 |
| Image recording | Category: CCD / Film or detector model: GENERIC TVIPS (4k x 4k) / Number real images: 200 / Average electron dose: 10 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Nitrogen cooled / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Details | The particles were submitted to single-particle analysis with icosahedral symmetry using IMAGIC-V |
|---|---|
| CTF correction | Details: Images |
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 13.0 Å / Resolution method: OTHER / Software - Name: IMAGIC / Number images used: 3329 |
| Final angle assignment | Details: ICOSAHEDRAL |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Protocol: Rigid body. Manually fitted 60 copies of the hexamer of HK97. Every hexamer was refined using Chimera. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 10 |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















