[English] 日本語
 Yorodumi
Yorodumi- EMDB-24393: Cryo-EM structure of human binary NatC complex with a Bisubstrate... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human binary NatC complex with a Bisubstrate inhibitor | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NatC / NAA30 / NAA35 / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationN-terminal methionine Nalpha-acetyltransferase NatC / NatC complex / smooth muscle cell proliferation / Retrograde transport at the Trans-Golgi-Network / peptide alpha-N-acetyltransferase activity / negative regulation of apoptotic process / nucleoplasm / nucleus / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Deng S / Marmorstein R | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: Molecular role of NAA38 in thermostability and catalytic activity of the human NatC N-terminal acetyltransferase. Authors: Sunbin Deng / Sarah M Gardner / Leah Gottlieb / Buyan Pan / E James Petersson / Ronen Marmorstein /  Abstract: N-terminal acetylation occurs on over 80% of human proteins and is catalyzed by a family of N-terminal acetyltransferases (NATs). All NATs contain a small catalytic subunit, while some also contain a ...N-terminal acetylation occurs on over 80% of human proteins and is catalyzed by a family of N-terminal acetyltransferases (NATs). All NATs contain a small catalytic subunit, while some also contain a large auxiliary subunit that facilitates catalysis and ribosome targeting for co-translational acetylation. NatC is one of the major NATs containing an NAA30 catalytic subunit, but uniquely contains two auxiliary subunits, large NAA35 and small NAA38. Here, we report the cryo-EM structures of human NatC (hNatC) complexes with and without NAA38, together with biochemical studies, to reveal that NAA38 increases the thermostability and broadens the substrate-specificity profile of NatC by ordering an N-terminal segment of NAA35 and reorienting an NAA30 N-terminal peptide binding loop for optimal catalysis, respectively. We also note important differences in engagement with a stabilizing inositol hexaphosphate molecule between human and yeast NatC. These studies provide new insights for the function and evolution of the NatC complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24393.map.gz emd_24393.map.gz | 28 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24393-v30.xml emd-24393-v30.xml emd-24393.xml emd-24393.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24393.png emd_24393.png | 102.1 KB | ||
| Filedesc metadata |  emd-24393.cif.gz emd-24393.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24393 http://ftp.pdbj.org/pub/emdb/structures/EMD-24393 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24393 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24393 | HTTPS FTP |
-Validation report
| Summary document |  emd_24393_validation.pdf.gz emd_24393_validation.pdf.gz | 528.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24393_full_validation.pdf.gz emd_24393_full_validation.pdf.gz | 527.7 KB | Display | |
| Data in XML |  emd_24393_validation.xml.gz emd_24393_validation.xml.gz | 5.8 KB | Display | |
| Data in CIF |  emd_24393_validation.cif.gz emd_24393_validation.cif.gz | 6.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24393 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24393 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24393 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24393 | HTTPS FTP |
-Related structure data
| Related structure data |  7rb3MC  7mx2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24393.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24393.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : heterotimeric S.pombe NatC complex with a bisubstrate inhibitor a...
| Entire | Name: heterotimeric S.pombe NatC complex with a bisubstrate inhibitor and inositol hexaphosphate |
|---|---|
| Components |
|
-Supramolecule #1: heterotimeric S.pombe NatC complex with a bisubstrate inhibitor a...
| Supramolecule | Name: heterotimeric S.pombe NatC complex with a bisubstrate inhibitor and inositol hexaphosphate type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: N-alpha-acetyltransferase 35, NatC auxiliary subunit
| Macromolecule | Name: N-alpha-acetyltransferase 35, NatC auxiliary subunit / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 80.707625 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: NWVDITQDFE EACRELKLGE LLHDKLFGLF EAMSAIEMMD PKMDAGMIGN QVNRKVLNFE QAIKDGTIKI KDLTLPELIG IMDTCFCCL ITWLEGHSLA QTVFTCLYIH NPDFIEDPAM KAFALGILKI CDIAREKVNK AAVFEEEDFQ SMTYGFKMAN S VTDLRVTG ...String: NWVDITQDFE EACRELKLGE LLHDKLFGLF EAMSAIEMMD PKMDAGMIGN QVNRKVLNFE QAIKDGTIKI KDLTLPELIG IMDTCFCCL ITWLEGHSLA QTVFTCLYIH NPDFIEDPAM KAFALGILKI CDIAREKVNK AAVFEEEDFQ SMTYGFKMAN S VTDLRVTG MLKDVEDDMQ RRVKSTRSRQ GEERDPEVEL EHQQCLAVFS RVKFTRVLLT VLIAFTKKET SAVAEAQKLM VQ AADLLSA IHNSLHHGIQ AQNDTTKGDH PIMMGFEPLV NQRLLPPTFP RYAKIIKREE MVNYFARLID RIKTVCEVVN LTN LHCILD FFCEFSEQSP CVLSRSLLQT TFLVDNKKVF GTHLMQDMVK DALRSFVSPP VLSPKCYLYN NHQAKDCIDS FVTH CVRPF CSLIQIHGHN RARQRDKLGH ILEEFATLQD EAEKVDAALH TMLLKQEPQR QHLACLGTWV LYHNLRIMIQ YLLSG FELE LYSMHEYYYI YWYLSEFLYA WLMSTLSRAD GSQMAEERIM EEQQKGRSSK KTKKKKKVRP LSREITMSQA YQNMCA GMF KTMVAFDMDG KVRKPKFELD SEQVRYEHRF APFNSVMTPP PVHYLQFKEM SDLNKYSPPP QSPELYVAAS KHFQQAK MI LENIPNPDHE VNRILKVAKP NFVVMKLLAG GHKKESKVPP EFDFSAHKYF PVVKLV UniProtKB: N-alpha-acetyltransferase 35, NatC auxiliary subunit |
-Macromolecule #2: N-alpha-acetyltransferase 30
| Macromolecule | Name: N-alpha-acetyltransferase 30 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number: N-terminal methionine Nalpha-acetyltransferase NatC |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 18.083293 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: RTIRYVRYES ELQMPDIMRL ITKDLSEPYS IYTYRYFIHN WPQLCFLAMV GEECVGAIVC KLDMHKKMFR RGYIAMLAVD SKYRRNGIG TNLVKKAIYA MVEGDCDEVV LETEITNKSA LKLYENLGFV RDKRLFRYYL NGVDALRLKL WLR UniProtKB: N-alpha-acetyltransferase 30 |
-Macromolecule #3: CARBOXYMETHYL COENZYME *A
| Macromolecule | Name: CARBOXYMETHYL COENZYME *A / type: ligand / ID: 3 / Number of copies: 1 / Formula: CMC |
|---|---|
| Molecular weight | Theoretical: 825.57 Da |
| Chemical component information | 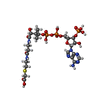 ChemComp-CMC: |
-Macromolecule #4: METHIONINE
| Macromolecule | Name: METHIONINE / type: ligand / ID: 4 / Number of copies: 1 / Formula: MET |
|---|---|
| Molecular weight | Theoretical: 149.211 Da |
| Chemical component information |  ChemComp-MET: |
-Macromolecule #5: LEUCINE
| Macromolecule | Name: LEUCINE / type: ligand / ID: 5 / Number of copies: 1 / Formula: LEU |
|---|---|
| Molecular weight | Theoretical: 131.173 Da |
| Chemical component information |  ChemComp-LEU: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 289 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K3 (6k x 4k) / #0 - Average electron dose: 1.6 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K3 (6k x 4k) / #1 - Average electron dose: 1.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7rb3: |
 Movie
Movie Controller
Controller