+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24265 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
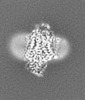
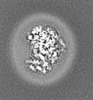
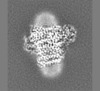
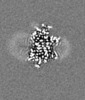
| Title | E. coli cytochrome bo3 in MSP nanodisc | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ubiquinone / Heme-copper Oxidoreductase / Electron transport / Bioenergetics / Proton pump / Membrane protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationcytochrome bo3 ubiquinol oxidase activity => GO:0009486 / ubiquinol oxidase (H+-transporting) / cytochrome bo3 ubiquinol oxidase activity / aerobic electron transport chain / oxidoreductase activity, acting on diphenols and related substances as donors, oxygen as acceptor / cytochrome-c oxidase activity / electron transport coupled proton transport / ATP synthesis coupled electron transport / membrane => GO:0016020 / : ...cytochrome bo3 ubiquinol oxidase activity => GO:0009486 / ubiquinol oxidase (H+-transporting) / cytochrome bo3 ubiquinol oxidase activity / aerobic electron transport chain / oxidoreductase activity, acting on diphenols and related substances as donors, oxygen as acceptor / cytochrome-c oxidase activity / electron transport coupled proton transport / ATP synthesis coupled electron transport / membrane => GO:0016020 / : / aerobic respiration / copper ion binding / heme binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.19 Å | |||||||||
 Authors Authors | Vallese F / Clarke OB | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Cryo-EM structures of cytochrome reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site. Authors: Jiao Li / Long Han / Francesca Vallese / Ziqiao Ding / Sylvia K Choi / Sangjin Hong / Yanmei Luo / Bin Liu / Chun Kit Chan / Emad Tajkhorshid / Jiapeng Zhu / Oliver Clarke / Kai Zhang / Robert Gennis /   Abstract: Two independent structures of the proton-pumping, respiratory cytochrome ubiquinol oxidase (cyt ) have been determined by cryogenic electron microscopy (cryo-EM) in styrene-maleic acid (SMA) ...Two independent structures of the proton-pumping, respiratory cytochrome ubiquinol oxidase (cyt ) have been determined by cryogenic electron microscopy (cryo-EM) in styrene-maleic acid (SMA) copolymer nanodiscs and in membrane scaffold protein (MSP) nanodiscs to 2.55- and 2.19-Å resolution, respectively. The structures include the metal redox centers (heme , heme , and Cu), the redox-active cross-linked histidine-tyrosine cofactor, and the internal water molecules in the proton-conducting D channel. Each structure also contains one equivalent of ubiquinone-8 (UQ8) in the substrate binding site as well as several phospholipid molecules. The isoprene side chain of UQ8 is clamped within a hydrophobic groove in subunit I by transmembrane helix TM0, which is only present in quinol oxidases and not in the closely related cytochrome oxidases. Both structures show carbonyl O1 of the UQ8 headgroup hydrogen bonded to D75 and R71 In both structures, residue H98 occupies two conformations. In conformation 1, H98 forms a hydrogen bond with carbonyl O4 of the UQ8 headgroup, but in conformation 2, the imidazole side chain of H98 has flipped to form a hydrogen bond with E14 at the N-terminal end of TM0. We propose that H98 dynamics facilitate proton transfer from ubiquinol to the periplasmic aqueous phase during oxidation of the substrate. Computational studies show that TM0 creates a channel, allowing access of water to the ubiquinol headgroup and to H98. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24265.map.gz emd_24265.map.gz | 242 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24265-v30.xml emd-24265-v30.xml emd-24265.xml emd-24265.xml | 24.8 KB 24.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24265.png emd_24265.png | 412.6 KB | ||
| Filedesc metadata |  emd-24265.cif.gz emd-24265.cif.gz | 7.1 KB | ||
| Others |  emd_24265_additional_1.map.gz emd_24265_additional_1.map.gz emd_24265_half_map_1.map.gz emd_24265_half_map_1.map.gz emd_24265_half_map_2.map.gz emd_24265_half_map_2.map.gz | 241.2 MB 475.8 MB 475.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24265 http://ftp.pdbj.org/pub/emdb/structures/EMD-24265 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24265 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24265 | HTTPS FTP |
-Validation report
| Summary document |  emd_24265_validation.pdf.gz emd_24265_validation.pdf.gz | 918.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24265_full_validation.pdf.gz emd_24265_full_validation.pdf.gz | 917.7 KB | Display | |
| Data in XML |  emd_24265_validation.xml.gz emd_24265_validation.xml.gz | 16.2 KB | Display | |
| Data in CIF |  emd_24265_validation.cif.gz emd_24265_validation.cif.gz | 18.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24265 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24265 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24265 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24265 | HTTPS FTP |
-Related structure data
| Related structure data |  7n9zMC  7cubC  7cuqC  7cuwC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10737 (Title: Cryo-EM structures of E. coli cytochrome bo3 in MSP Nanodiscs EMPIAR-10737 (Title: Cryo-EM structures of E. coli cytochrome bo3 in MSP NanodiscsData size: 851.4 Data #1: Unaligned multi-frame micrographs of cyt bo3 in MSP Nanodiscs [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24265.map.gz / Format: CCP4 / Size: 260.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24265.map.gz / Format: CCP4 / Size: 260.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.41385 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: #1
| File | emd_24265_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
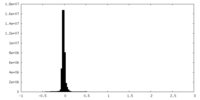
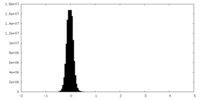
| Density Histograms |
-Half map: #2
| File | emd_24265_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
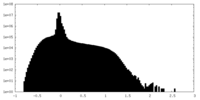
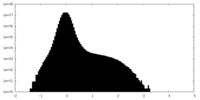
| Density Histograms |
-Half map: #1
| File | emd_24265_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
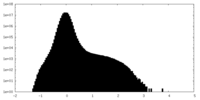
| Density Histograms |
- Sample components
Sample components
+Entire : Cytochrome bo 3 ubiquinol oxidase
+Supramolecule #1: Cytochrome bo 3 ubiquinol oxidase
+Macromolecule #1: Cytochrome o ubiquinol oxidase, subunit I
+Macromolecule #2: Ubiquinol oxidase subunit 2
+Macromolecule #3: Cytochrome o ubiquinol oxidase
+Macromolecule #4: Cytochrome o ubiquinol oxidase, subunit IV
+Macromolecule #5: Ubiquinone-8
+Macromolecule #6: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
+Macromolecule #7: 1,2-Distearoyl-sn-glycerophosphoethanolamine
+Macromolecule #8: HEME O
+Macromolecule #9: PROTOPORPHYRIN IX CONTAINING FE
+Macromolecule #10: CARDIOLIPIN
+Macromolecule #11: COPPER (II) ION
+Macromolecule #12: ZINC ION
+Macromolecule #13: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.6 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 / Component:
| ||||||
| Grid | Model: UltrAuFoil R0.6/1 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 58.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.19 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 94681 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)