+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24090 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of minimal TRPV1 with RTX bound in C2 state | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | TRP channel / cryo-EM / nanodisc / vanilloid agonist / TRANSPORT PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationtemperature-gated ion channel activity / response to capsazepine / negative regulation of establishment of blood-brain barrier / sensory perception of mechanical stimulus / peptide secretion / urinary bladder smooth muscle contraction / detection of chemical stimulus involved in sensory perception of pain / smooth muscle contraction involved in micturition / TRP channels / cellular response to temperature stimulus ...temperature-gated ion channel activity / response to capsazepine / negative regulation of establishment of blood-brain barrier / sensory perception of mechanical stimulus / peptide secretion / urinary bladder smooth muscle contraction / detection of chemical stimulus involved in sensory perception of pain / smooth muscle contraction involved in micturition / TRP channels / cellular response to temperature stimulus / cellular response to acidic pH / fever generation / detection of temperature stimulus involved in thermoception / thermoception / negative regulation of systemic arterial blood pressure / glutamate secretion / response to pH / chloride channel regulator activity / dendritic spine membrane / monoatomic cation transmembrane transporter activity / excitatory extracellular ligand-gated monoatomic ion channel activity / negative regulation of heart rate / cellular response to ATP / temperature homeostasis / response to pain / cellular response to alkaloid / calcium ion import across plasma membrane / diet induced thermogenesis / cellular response to cytokine stimulus / detection of temperature stimulus involved in sensory perception of pain / behavioral response to pain / intracellularly gated calcium channel activity / negative regulation of mitochondrial membrane potential / ligand-gated monoatomic ion channel activity / extracellular ligand-gated monoatomic ion channel activity / monoatomic cation channel activity / GABA-ergic synapse / sensory perception of pain / phosphatidylinositol binding / cellular response to nerve growth factor stimulus / phosphoprotein binding / calcium ion transmembrane transport / microglial cell activation / calcium channel activity / lipid metabolic process / cellular response to growth factor stimulus / response to peptide hormone / calcium ion transport / positive regulation of nitric oxide biosynthetic process / transmembrane signaling receptor activity / cellular response to tumor necrosis factor / cellular response to heat / positive regulation of cytosolic calcium ion concentration / response to heat / monoatomic ion transmembrane transport / postsynaptic membrane / protein homotetramerization / calmodulin binding / neuron projection / positive regulation of apoptotic process / external side of plasma membrane / neuronal cell body / dendrite / negative regulation of transcription by RNA polymerase II / ATP binding / identical protein binding / membrane / metal ion binding / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | ||||||||||||||||||
 Authors Authors | Zhang K / Julius D | ||||||||||||||||||
| Funding support |  France, France,  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2021 Journal: Cell / Year: 2021Title: Structural snapshots of TRPV1 reveal mechanism of polymodal functionality. Authors: Kaihua Zhang / David Julius / Yifan Cheng /  Abstract: Many transient receptor potential (TRP) channels respond to diverse stimuli and conditionally conduct small and large cations. Such functional plasticity is presumably enabled by a uniquely dynamic ...Many transient receptor potential (TRP) channels respond to diverse stimuli and conditionally conduct small and large cations. Such functional plasticity is presumably enabled by a uniquely dynamic ion selectivity filter that is regulated by physiological agents. What is currently missing is a "photo series" of intermediate structural states that directly address this hypothesis and reveal specific mechanisms behind such dynamic channel regulation. Here, we exploit cryoelectron microscopy (cryo-EM) to visualize conformational transitions of the capsaicin receptor, TRPV1, as a model to understand how dynamic transitions of the selectivity filter in response to algogenic agents, including protons, vanilloid agonists, and peptide toxins, permit permeation by small and large organic cations. These structures also reveal mechanisms governing ligand binding substates, as well as allosteric coupling between key sites that are proximal to the selectivity filter and cytoplasmic gate. These insights suggest a general framework for understanding how TRP channels function as polymodal signal integrators. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24090.map.gz emd_24090.map.gz | 117 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24090-v30.xml emd-24090-v30.xml emd-24090.xml emd-24090.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24090_fsc.xml emd_24090_fsc.xml | 10.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_24090.png emd_24090.png | 177.1 KB | ||
| Filedesc metadata |  emd-24090.cif.gz emd-24090.cif.gz | 5.6 KB | ||
| Others |  emd_24090_half_map_1.map.gz emd_24090_half_map_1.map.gz emd_24090_half_map_2.map.gz emd_24090_half_map_2.map.gz | 93.7 MB 94.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24090 http://ftp.pdbj.org/pub/emdb/structures/EMD-24090 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24090 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24090 | HTTPS FTP |
-Validation report
| Summary document |  emd_24090_validation.pdf.gz emd_24090_validation.pdf.gz | 889.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24090_full_validation.pdf.gz emd_24090_full_validation.pdf.gz | 888.7 KB | Display | |
| Data in XML |  emd_24090_validation.xml.gz emd_24090_validation.xml.gz | 18 KB | Display | |
| Data in CIF |  emd_24090_validation.cif.gz emd_24090_validation.cif.gz | 22.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24090 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24090 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24090 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24090 | HTTPS FTP |
-Related structure data
| Related structure data |  7mzdMC  7l2hC  7l2iC  7l2jC  7l2kC  7l2lC  7l2mC  7l2nC  7l2oC  7l2pC  7l2rC  7l2sC  7l2tC  7l2uC  7l2vC  7l2wC  7l2xC  7mz5C  7mz6C  7mz7C  7mz9C  7mzaC  7mzbC  7mzcC  7mzeC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24090.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24090.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.834 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
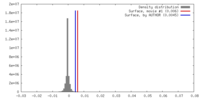
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_24090_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
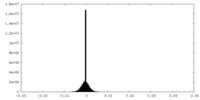
| Density Histograms |
-Half map: #2
| File | emd_24090_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
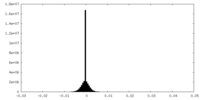
| Density Histograms |
- Sample components
Sample components
-Entire : minimal TRPV1 in nanodisc
| Entire | Name: minimal TRPV1 in nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: minimal TRPV1 in nanodisc
| Supramolecule | Name: minimal TRPV1 in nanodisc / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Transient receptor potential cation channel subfamily V member 1
| Macromolecule | Name: Transient receptor potential cation channel subfamily V member 1 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 73.016352 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GAMGSRLYDR RSIFDAVAQS NCQELESLLP FLQRSKKRLT DSEFKDPETG KTCLLKAMLN LHNGQNDTIA LLLDVARKTD SLKQFVNAS YTDSYYKGQT ALHIAIERRN MTLVTLLVEN GADVQAAANG DFFKKTKGRP GFYFGELPLS LAACTNQLAI V KFLLQNSW ...String: GAMGSRLYDR RSIFDAVAQS NCQELESLLP FLQRSKKRLT DSEFKDPETG KTCLLKAMLN LHNGQNDTIA LLLDVARKTD SLKQFVNAS YTDSYYKGQT ALHIAIERRN MTLVTLLVEN GADVQAAANG DFFKKTKGRP GFYFGELPLS LAACTNQLAI V KFLLQNSW QPADISARDS VGNTVLHALV EVADNTVDNT KFVTSMYNEI LILGAKLHPT LKLEEITNRK GLTPLALAAS SG KIGVLAY ILQREIHEPE CRHLSRKFTE WAYGPVHSSL YDLSCIDTCE KNSVLEVIAY SSSETPNRHD MLLVEPLNRL LQD KWDRFV KRIFYFNFFV YCLYMIIFTA AAYYRPVEGL PPYKLKNTVG DYFRVTGEIL SVSGGVYFFF RGIQYFLQRR PSLK SLFVD SYSEILFFVQ SLFMLVSVVL YFSQRKEYVA SMVFSLAMGW TNMLYYTRGF QQMGIYAVMI EKMILRDLCR FMFVY LVFL FGFSTAVVTL IEDGKYNSLY STCLELFKFT IGMGDLEFTE NYDFKAVFII LLLAYVILTY ILLLNMLIAL MGETVN KIA QESKNIWKLQ RAITILDTEK SFLKCMRKAF RSGKLLQVGF TPDGKDDYRW CFRVDEVNWT TWNTNVGIIN EDPG UniProtKB: Transient receptor potential cation channel subfamily V member 1 |
-Macromolecule #2: resiniferatoxin
| Macromolecule | Name: resiniferatoxin / type: ligand / ID: 2 / Number of copies: 4 / Formula: 6EU |
|---|---|
| Molecular weight | Theoretical: 628.708 Da |
| Chemical component information |  ChemComp-6EU: |
-Macromolecule #3: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 3 / Number of copies: 2 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 4 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.3 |
|---|---|
| Sugar embedding | Material: nanodisc |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)