+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24006 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Legionella pneumophila Dot/Icm T4SS PR | |||||||||||||||
 Map data Map data | Reconstruction of the Legionella pneumophila Dot/Icm T4SS PR | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Dot/Icm / Secretion / T4SS / TRANSLOCASE | |||||||||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||||||||
 Authors Authors | Sheedlo MJ / Durie CL | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: Cryo-EM reveals new species-specific proteins and symmetry elements in the Dot/Icm T4SS. Authors: Michael J Sheedlo / Clarissa L Durie / Jeong Min Chung / Louise Chang / Jacquelyn Roberts / Michele Swanson / Dana Borden Lacy / Melanie D Ohi /   Abstract: is an opportunistic pathogen that causes the potentially fatal pneumonia known as Legionnaires' disease. The pathology associated with infection depends on bacterial delivery of effector proteins ... is an opportunistic pathogen that causes the potentially fatal pneumonia known as Legionnaires' disease. The pathology associated with infection depends on bacterial delivery of effector proteins into the host via the membrane spanning Dot/Icm type IV secretion system (T4SS). We have determined sub-3.0 Å resolution maps of the Dot/Icm T4SS core complex by single particle cryo-EM. The high-resolution structural analysis has allowed us to identify proteins encoded outside the Dot/Icm genetic locus that contribute to the core T4SS structure. We can also now define two distinct areas of symmetry mismatch, one that connects the C18 periplasmic ring (PR) and the C13 outer membrane cap (OMC) and one that connects the C13 OMC with a 16-fold symmetric dome. Unexpectedly, the connection between the PR and OMC is DotH, with five copies sandwiched between the OMC and PR to accommodate the symmetry mismatch. Finally, we observe multiple conformations in the reconstructions that indicate flexibility within the structure. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24006.map.gz emd_24006.map.gz | 23.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24006-v30.xml emd-24006-v30.xml emd-24006.xml emd-24006.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24006.png emd_24006.png | 62.7 KB | ||
| Filedesc metadata |  emd-24006.cif.gz emd-24006.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24006 http://ftp.pdbj.org/pub/emdb/structures/EMD-24006 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24006 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24006 | HTTPS FTP |
-Validation report
| Summary document |  emd_24006_validation.pdf.gz emd_24006_validation.pdf.gz | 340.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24006_full_validation.pdf.gz emd_24006_full_validation.pdf.gz | 340.4 KB | Display | |
| Data in XML |  emd_24006_validation.xml.gz emd_24006_validation.xml.gz | 7.8 KB | Display | |
| Data in CIF |  emd_24006_validation.cif.gz emd_24006_validation.cif.gz | 9.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24006 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24006 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24006 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24006 | HTTPS FTP |
-Related structure data
| Related structure data |  7mueMC  7mucC  7mudC  7muqC  7musC  7muvC  7muwC  7muyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24006.map.gz / Format: CCP4 / Size: 506 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24006.map.gz / Format: CCP4 / Size: 506 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of the Legionella pneumophila Dot/Icm T4SS PR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
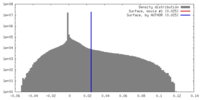
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Legionella pneumophila Dot/Icm T4SS PR
| Entire | Name: Legionella pneumophila Dot/Icm T4SS PR |
|---|---|
| Components |
|
-Supramolecule #1: Legionella pneumophila Dot/Icm T4SS PR
| Supramolecule | Name: Legionella pneumophila Dot/Icm T4SS PR / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: DotF
| Macromolecule | Name: DotF / type: protein_or_peptide / ID: 1 / Number of copies: 18 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 29.729969 KDa |
| Sequence | String: MMAEHDQNND EYKFAELDSY DMDQAGESDL DSEASYQSGK EGLTKKKDIK RNALIAIGAV VFIMVMYKII GWMFFSDKSS QVTSKPAIP PVTQVATPQP VQTIPTTTPI QQVQPTTIIE DDPDLKKKVS AIEMTQQSLR SEVNALSEQI NAVNNNIKNL N AQIVNLNQ ...String: MMAEHDQNND EYKFAELDSY DMDQAGESDL DSEASYQSGK EGLTKKKDIK RNALIAIGAV VFIMVMYKII GWMFFSDKSS QVTSKPAIP PVTQVATPQP VQTIPTTTPI QQVQPTTIIE DDPDLKKKVS AIEMTQQSLR SEVNALSEQI NAVNNNIKNL N AQIVNLNQ IIGNMSNQIA RQSEVINVLM ARTTPKKVVK VSRPIVQARI IYYIQAVIPG RAWLIGSNGS TLTVREGSKI PG YGMVKLI DSLQGRILTS SGQVIKFSQE DS UniProtKB: DotF |
-Macromolecule #2: Type IV secretion protein IcmK
| Macromolecule | Name: Type IV secretion protein IcmK / type: protein_or_peptide / ID: 2 / Number of copies: 18 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.958926 KDa |
| Sequence | String: MMKKYDQLCK YCLVIGLTFS MSCSIYAADQ SDDAQQALQQ LRMLQQKLSQ NPSPDAQSGA GDGGDNAASD STQQPNQSGQ ANAPAANQT ATAGGDGQII SQDDAEVIDK KAFKDMTRNL YPLNPEQVVK LKQIYETSEY AKAATPGTPP KPTATSQFVN L SPGSTPPV ...String: MMKKYDQLCK YCLVIGLTFS MSCSIYAADQ SDDAQQALQQ LRMLQQKLSQ NPSPDAQSGA GDGGDNAASD STQQPNQSGQ ANAPAANQT ATAGGDGQII SQDDAEVIDK KAFKDMTRNL YPLNPEQVVK LKQIYETSEY AKAATPGTPP KPTATSQFVN L SPGSTPPV IRLSQGFVSS LVFLDSTGAP WPIAAYDLGD PSSFNIQWDK TSNTLMIQAT KLYNYGNLAV RLRGLNTPVM LT LIPGQKA VDYRVDLRVQ GYGPNAKSMP TEEGIPPSAN DLLLHVLEGV PPPGSRRLVV SGGDARAWLS NEKMYVRTNL TIL SPGWLA SMTSADGTHA YEMQKSPVLL VSWHGKVMQL KVEGL UniProtKB: Type IV secretion protein IcmK |
-Macromolecule #3: Unknown protein fragment
| Macromolecule | Name: Unknown protein fragment / type: protein_or_peptide / ID: 3 / Number of copies: 18 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 4.103049 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #4: IcmE protein
| Macromolecule | Name: IcmE protein / type: protein_or_peptide / ID: 4 / Number of copies: 18 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 107.911891 KDa |
| Sequence | String: MASKKENLKS LFSNTRTRVI IIFTAALLII AVVIGFFKIR GATTGSIAAA EVSTVPGGIQ SIPGVLDPTA QYAKLQEEQN ITQAQVAEK TGGSAIPTII RTQALGEGVG VIGSQSGVGF AALAQEELGG PQRSLWIQEL QDGSCSKSVI TKVVNQGAQL T DLKAACSC ...String: MASKKENLKS LFSNTRTRVI IIFTAALLII AVVIGFFKIR GATTGSIAAA EVSTVPGGIQ SIPGVLDPTA QYAKLQEEQN ITQAQVAEK TGGSAIPTII RTQALGEGVG VIGSQSGVGF AALAQEELGG PQRSLWIQEL QDGSCSKSVI TKVVNQGAQL T DLKAACSC VQLKDSGYGL QELEQVCECK ELKSAGYNAR QLKEAGYSAG RLRNCGFDAC ELRNAGFTAQ EMKDGGFSDG EL KGAGFSD AEIAKASGLP DGITADDVRK AGCGAAALAK LRQAGVSASA IRKISGCTAE QLKAAGYTAK ELKDAGFSAA DLR RAGFSA AELKDAGFTA RDLLNAGFTP ADLAKAGFSD AQIKAAQAEL PPGITPQDVK NAGCDVEALK KEREAGVSAA LIRQ YAGCS AQALKAAGFT DADLANAGFT PAQISAATPL SDAEIKAAGC DPDKLKKLFS AGVSAKRIKE LNGCSAEALK AAGYD AQSL LAAGFTPQEL LAAGFTPKQL EDAGLNPVSI IADGRVADCS VESLKKARAA GVSALTIKQT LGCSAAALKA AGYTAK ELK DAGFTAAELK AAGFSAKELK DAGFTAKELR DAGFSAQELK DVGFSAKDLK DAGFSAAELK AAGFTAAQLK AAGFSAK DL KDAGFSAAEL KAAGFSAKEL KDAGFSASDL KNAGFSAKEL KDAGFSASDL KSAGFSASEL KNAGYSADEL KKAGYTSA E LRNAGFSPQE SAVAGLQGPD LQQLDSSITG IPSIPGATPR PTTSDAASSA EQLQAILQKQ NEQLAEQKYQ QEIQQRTSD MLTAATQLVQ DWKQVETQVY TEGTEETKTS GGESAVPGTG TGTGSNNQPV DQGAVSAQNQ AIIKTGDIMF AVLDTSVNSD EPGPILATI VTGKLKGSKL IGSFNLPSNA DKMVITFNTM SIPGAEKTIS ISAYAIDPNT ARTALASRTN HHYLMRYGSL F ASSFLQGF GNAFQSANTT ITIGGTGGGN NITVANGVGR STLENAVIGL ATVGKAWSQQ AQQLFNTPTT VEVYSGTGLG IL FTQDVTT I UniProtKB: IcmE protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 43907 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7mue: |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)