+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23608 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | DpK2 bacteriophage tail spike depolymerase | |||||||||
 Map data Map data | postprocessed map, C3 point group symmetry | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bateriophage tail spike / depolymerase / klebsiella targeting / SUGAR BINDING PROTEIN | |||||||||
| Function / homology | biological process involved in interaction with host / Pectin lyase fold/virulence factor / viral life cycle / virion component / Depolymerase Function and homology information Function and homology information | |||||||||
| Biological species |  Bacteriophage sp. (virus) / Bacteriophage sp. (virus) /  Klebsiella phage GH-K3 (virus) Klebsiella phage GH-K3 (virus) | |||||||||
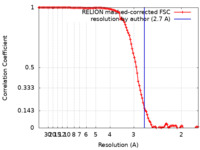
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Belousoff MJ / Bamert RS | |||||||||
| Funding support |  Australia, 1 items Australia, 1 items
| |||||||||
 Citation Citation |  Journal: Microbiol Spectr / Year: 2021 Journal: Microbiol Spectr / Year: 2021Title: Mechanistic Insights into the Capsule-Targeting Depolymerase from a Klebsiella pneumoniae Bacteriophage. Authors: Rhys A Dunstan / Rebecca S Bamert / Matthew J Belousoff / Francesca L Short / Christopher K Barlow / Derek J Pickard / Jonathan J Wilksch / Ralf B Schittenhelm / Richard A Strugnell / Gordon ...Authors: Rhys A Dunstan / Rebecca S Bamert / Matthew J Belousoff / Francesca L Short / Christopher K Barlow / Derek J Pickard / Jonathan J Wilksch / Ralf B Schittenhelm / Richard A Strugnell / Gordon Dougan / Trevor Lithgow /   Abstract: The production of capsular polysaccharides by Klebsiella pneumoniae protects the bacterial cell from harmful environmental factors such as antimicrobial compounds and infection by bacteriophages ...The production of capsular polysaccharides by Klebsiella pneumoniae protects the bacterial cell from harmful environmental factors such as antimicrobial compounds and infection by bacteriophages (phages). To bypass this protective barrier, some phages encode polysaccharide-degrading enzymes referred to as depolymerases to provide access to cell surface receptors. Here, we characterized the phage RAD2, which infects K. pneumoniae strains that produce the widespread, hypervirulence-associated K2-type capsular polysaccharide. Using transposon-directed insertion sequencing, we have shown that the production of capsule is an absolute requirement for efficient RAD2 infection by serving as a first-stage receptor. We have identified the depolymerase responsible for recognition and degradation of the capsule, determined that the depolymerase forms globular appendages on the phage virion tail tip, and present the cryo-electron microscopy structure of the RAD2 capsule depolymerase at 2.7-Å resolution. A putative active site for the enzyme was identified, comprising clustered negatively charged residues that could facilitate the hydrolysis of target polysaccharides. Enzymatic assays coupled with mass spectrometric analyses of digested oligosaccharide products provided further mechanistic insight into the hydrolase activity of the enzyme, which, when incubated with K. pneumoniae, removes the capsule and sensitizes the cells to serum-induced killing. Overall, these findings expand our understanding of how phages target the Klebsiella capsule for infection, providing a framework for the use of depolymerases as antivirulence agents against this medically important pathogen. Klebsiella pneumoniae is a medically important pathogen that produces a thick protective capsule that is essential for pathogenicity. Phages are natural predators of bacteria, and many encode diverse "capsule depolymerases" which specifically degrade the capsule of their hosts, an exploitable trait for potential therapies. We have determined the first structure of a depolymerase that targets the clinically relevant K2 capsule and have identified its putative active site, providing hints to its mechanism of action. We also show that Klebsiella cells treated with a recombinant form of the depolymerase are stripped of capsule, inhibiting their ability to grow in the presence of serum, demonstrating the anti-infective potential of these robust and readily producible enzymes against encapsulated bacterial pathogens such as K. pneumoniae. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23608.map.gz emd_23608.map.gz | 117.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23608-v30.xml emd-23608-v30.xml emd-23608.xml emd-23608.xml | 22.9 KB 22.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23608_fsc.xml emd_23608_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_23608.png emd_23608.png | 59.2 KB | ||
| Filedesc metadata |  emd-23608.cif.gz emd-23608.cif.gz | 6 KB | ||
| Others |  emd_23608_additional_1.map.gz emd_23608_additional_1.map.gz emd_23608_additional_2.map.gz emd_23608_additional_2.map.gz emd_23608_additional_3.map.gz emd_23608_additional_3.map.gz emd_23608_additional_4.map.gz emd_23608_additional_4.map.gz emd_23608_half_map_1.map.gz emd_23608_half_map_1.map.gz emd_23608_half_map_2.map.gz emd_23608_half_map_2.map.gz | 78.3 MB 98.3 MB 117.3 MB 98.6 MB 99 MB 99 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23608 http://ftp.pdbj.org/pub/emdb/structures/EMD-23608 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23608 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23608 | HTTPS FTP |
-Validation report
| Summary document |  emd_23608_validation.pdf.gz emd_23608_validation.pdf.gz | 786.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23608_full_validation.pdf.gz emd_23608_full_validation.pdf.gz | 786 KB | Display | |
| Data in XML |  emd_23608_validation.xml.gz emd_23608_validation.xml.gz | 18.6 KB | Display | |
| Data in CIF |  emd_23608_validation.cif.gz emd_23608_validation.cif.gz | 24.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23608 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23608 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23608 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23608 | HTTPS FTP |
-Related structure data
| Related structure data |  7lzjMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_23608.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23608.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | postprocessed map, C3 point group symmetry | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.895 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: local resolution filtered map, C3 point group symmetry
| File | emd_23608_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local resolution filtered map, C3 point group symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
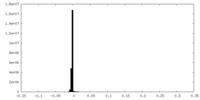
| Density Histograms |
-Additional map: unprocessed map, C3 point group symmetry
| File | emd_23608_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unprocessed map, C3 point group symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
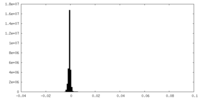
| Density Histograms |
-Additional map: post-processed map, C1 point group symmetry
| File | emd_23608_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | post-processed map, C1 point group symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
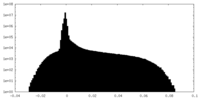
| Density Histograms |
-Additional map: unprocessed map, C1 point group symmetry
| File | emd_23608_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unprocessed map, C1 point group symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
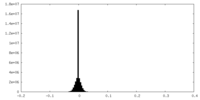
| Density Histograms |
-Half map: half map 1, C3 point group symmetry
| File | emd_23608_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1, C3 point group symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2, C3 point group symmetry
| File | emd_23608_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2, C3 point group symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : DpK2 depolymerase tail spike complex
| Entire | Name: DpK2 depolymerase tail spike complex |
|---|---|
| Components |
|
-Supramolecule #1: DpK2 depolymerase tail spike complex
| Supramolecule | Name: DpK2 depolymerase tail spike complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Bacteriophage sp. (virus) Bacteriophage sp. (virus) |
-Macromolecule #1: Depolymerase
| Macromolecule | Name: Depolymerase / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Klebsiella phage GH-K3 (virus) Klebsiella phage GH-K3 (virus) |
| Molecular weight | Theoretical: 98.437008 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MALYREGKAA MAADGTVTGT GTKWQSSLSL IRPGATIMFL SSPIQMAVVN KVVSDTEIKA ITTSGAVVAS TDYAILLSDS LTVDGLAQD VAETLRHYQS QETVIADAVE FFKSFDFDSL QNLANQIKAD SESAESSAAA AAASESKAKT SEDNAKSSEN A AKNSEVAA ...String: MALYREGKAA MAADGTVTGT GTKWQSSLSL IRPGATIMFL SSPIQMAVVN KVVSDTEIKA ITTSGAVVAS TDYAILLSDS LTVDGLAQD VAETLRHYQS QETVIADAVE FFKSFDFDSL QNLANQIKAD SESAESSAAA AAASESKAKT SEDNAKSSEN A AKNSEVAA ETTRDQIQQI IDNAGDQSTL VVLAQPDGFD SIGRVSSFAA LRNLKPKKSG QHVLLTSYYD GWAAENKMPT GG GEFISSI GTATDDGGYI AAGPGYYWTR VVNNNSFTAE DFGCKTTATP PPNFNVLPAE LFDNTAMMQA AFNLAISKSF KLN LSTGTY YFESSDTLRI TGPIHIEGRP GTVFYHNPSN KANPKTDAFM NISGCSAGRI SSINCFSNSY LGKGINFDRS VGDN RKLVL EHVYVDTFRW GFYVGEPECI NQIEFHSCRA QSNYFQGIFI ESFKEGQEYG HSAPVHFFNT ICNGNGPTSF ALGAT YKTT KNEYIKVMDS VNDVGCQAYF QGLSNVQYIG GQLSGHGSPR NTSLATITQC NSFIIYGTDL EDINGFTTDG TAITAD NID AIESNYLKDI SGAAIVVSSC PGFKIDSPHI FKIKTLSTIK LMNNTYNYEI GGFTPDEALK YNVWDANGLA TNRISGV IH PRLVNSRLGI NSVAFDNMSN KLDVSSLIHN ETSQIVGLTP STGSNVPHTR KMWSNGAMYS STDLNNGFRL NYLSNHNE P LTPMHLYNEF SVSEFGGSVT ESNALDEIKY IFIQTTYANS GDGRFIIQAL DASGSVLSSN WYSPQSFNST FPISGFVRF DVPTGAKKIR YGFVNSANYT GSLRSHFMSG FAYNKRFFLK IYAVYNDLGR YGQFEPPYSV AIDRFRVGDN TTQMPSIPAS SATDVAGVN EVINSLLASL KANGFMSS UniProtKB: Depolymerase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 43.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)