+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23514 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Proximal Ku in NHEJ Short-range synaptic complex | |||||||||||||||||||||
 Map data Map data | Proximal Ku in NHEJ Short-range synaptic complex | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.9 Å | |||||||||||||||||||||
 Authors Authors | He Y / Chen S | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Structural basis of long-range to short-range synaptic transition in NHEJ. Authors: Siyu Chen / Linda Lee / Tasmin Naila / Susan Fishbain / Annie Wang / Alan E Tomkinson / Susan P Lees-Miller / Yuan He /   Abstract: DNA double-strand breaks (DSBs) are a highly cytotoxic form of DNA damage and the incorrect repair of DSBs is linked to carcinogenesis. The conserved error-prone non-homologous end joining (NHEJ) ...DNA double-strand breaks (DSBs) are a highly cytotoxic form of DNA damage and the incorrect repair of DSBs is linked to carcinogenesis. The conserved error-prone non-homologous end joining (NHEJ) pathway has a key role in determining the effects of DSB-inducing agents that are used to treat cancer as well as the generation of the diversity in antibodies and T cell receptors. Here we applied single-particle cryo-electron microscopy to visualize two key DNA-protein complexes that are formed by human NHEJ factors. The Ku70/80 heterodimer (Ku), the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs), DNA ligase IV (LigIV), XRCC4 and XLF form a long-range synaptic complex, in which the DNA ends are held approximately 115 Å apart. Two DNA end-bound subcomplexes comprising Ku and DNA-PKcs are linked by interactions between the DNA-PKcs subunits and a scaffold comprising LigIV, XRCC4, XLF, XRCC4 and LigIV. The relative orientation of the DNA-PKcs molecules suggests a mechanism for autophosphorylation in trans, which leads to the dissociation of DNA-PKcs and the transition into the short-range synaptic complex. Within this complex, the Ku-bound DNA ends are aligned for processing and ligation by the XLF-anchored scaffold, and a single catalytic domain of LigIV is stably associated with a nick between the two Ku molecules, which suggests that the joining of both strands of a DSB involves both LigIV molecules. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23514.map.gz emd_23514.map.gz | 22.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23514-v30.xml emd-23514-v30.xml emd-23514.xml emd-23514.xml | 19.9 KB 19.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23514_fsc.xml emd_23514_fsc.xml | 6.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_23514.png emd_23514.png | 91.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23514 http://ftp.pdbj.org/pub/emdb/structures/EMD-23514 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23514 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23514 | HTTPS FTP |
-Validation report
| Summary document |  emd_23514_validation.pdf.gz emd_23514_validation.pdf.gz | 322.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23514_full_validation.pdf.gz emd_23514_full_validation.pdf.gz | 321.7 KB | Display | |
| Data in XML |  emd_23514_validation.xml.gz emd_23514_validation.xml.gz | 9.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23514 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23514 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23514 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23514 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_23514.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23514.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Proximal Ku in NHEJ Short-range synaptic complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.66 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
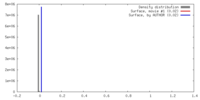
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Proximal Ku in NHEJ Short-range synaptic complex
| Entire | Name: Proximal Ku in NHEJ Short-range synaptic complex |
|---|---|
| Components |
|
-Supramolecule #1: Proximal Ku in NHEJ Short-range synaptic complex
| Supramolecule | Name: Proximal Ku in NHEJ Short-range synaptic complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Ku |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 150 KDa |
-Macromolecule #1: XRCC6_HUMAN X-ray repair cross-complementing protein 6
| Macromolecule | Name: XRCC6_HUMAN X-ray repair cross-complementing protein 6 type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: MSGWESYYKT EGDEEAEEEQ EENLEASGDY KYSGRDSLIF LVDASKAMFE SQSEDELTPF DMSIQCIQSV YISKIISSDR DLLAVVFYG TEKDKNSVNF KNIYVLQELD NPGAKRILEL DQFKGQQGQK RFQDMMGHGS DYSLSEVLWV CANLFSDVQF K MSHKRIML ...String: MSGWESYYKT EGDEEAEEEQ EENLEASGDY KYSGRDSLIF LVDASKAMFE SQSEDELTPF DMSIQCIQSV YISKIISSDR DLLAVVFYG TEKDKNSVNF KNIYVLQELD NPGAKRILEL DQFKGQQGQK RFQDMMGHGS DYSLSEVLWV CANLFSDVQF K MSHKRIML FTNEDNPHGN DSAKASRART KAGDLRDTGI FLDLMHLKKP GGFDISLFYR DIISIAEDED LRVHFEESSK LE DLLRKVR AKETRKRALS RLKLKLNKDI VISVGIYNLV QKALKPPPIK LYRETNEPVK TKTRTFNTST GGLLLPSDTK RSQ IYGSRQ IILEKEETEE LKRFDDPGLM LMGFKPLVLL KKHHYLRPSL FVYPEESLVI GSSTLFSALL IKCLEKEVAA LCRY TPRRN IPPYFVALVP QEEELDDQKI QVTPPGFQLV FLPFADDKRK MPFTEKIMAT PEQVGKMKAI VEKLRFTYRS DSFEN PVLQ QHFRNLEALA LDLMEPEQAV DLTLPKVEAM NKRLGSLVDE FKELVYPPDY NPEGKVTKRK HDNEGSGSKR PKVEYS EEE LKTHISKGTL GKFTVPMLKE ACRAYGLKSG LKKQELL |
-Macromolecule #2: XRCC5_HUMAN X-ray repair cross-complementing protein 5
| Macromolecule | Name: XRCC5_HUMAN X-ray repair cross-complementing protein 5 type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: MVRSGNKAAV VLCMDVGFTM SNSIPGIESP FEQAKKVITM FVQRQVFAEN KDEIALVLFG TDGTDNPLSG GDQYQNITVH RHLMLPDFD LLEDIESKIQ PGSQQADFLD ALIVSMDVIQ HETIGKKFEK RHIEIFTDLS SRFSKSQLDI IIHSLKKCDI S LQFFLPFS ...String: MVRSGNKAAV VLCMDVGFTM SNSIPGIESP FEQAKKVITM FVQRQVFAEN KDEIALVLFG TDGTDNPLSG GDQYQNITVH RHLMLPDFD LLEDIESKIQ PGSQQADFLD ALIVSMDVIQ HETIGKKFEK RHIEIFTDLS SRFSKSQLDI IIHSLKKCDI S LQFFLPFS LGKEDGSGDR GDGPFRLGGH GPSFPLKGIT EQQKEGLEIV KMVMISLEGE DGLDEIYSFS ESLRKLCVFK KI ERHSIHW PCRLTIGSNL SIRIAAYKSI LQERVKKTWT VVDAKTLKKE DIQKETVYCL NDDDETEVLK EDIIQGFRYG SDI VPFSKV DEEQMKYKSE GKCFSVLGFC KSSQVQRRFF MGNQVLKVFA ARDDEAAAVA LSSLIHALDD LDMVAIVRYA YDKR ANPQV GVAFPHIKHN YECLVYVQLP FMEDLRQYMF SSLKNSKKYA PTEAQLNAVD ALIDSMSLAK KDEKTDTLED LFPTT KIPN PRFQRLFQCL LHRALHPREP LPPIQQHIWN MLNPPAEVTT KSQIPLSKIK TLFPLIEAKK KDQVTAQEIF QDNHED GPT AKKLKTEQGG AHFSVSSLAE GSVTSVGSVN PAENFRVLVK QKKASFEEAS NQLINHIEQF LDTNETPYFM KSIDCIR AF REEAIKFSEE QRFNNFLKAL QEKVEIKQLN HFWEIVVQDG ITLITKEEAS GSSVTAEEAK KFLAPKDKPS GDTAAVFE E GGDVDDLLDM I |
-Macromolecule #3: DNA (5'-D(P*TP*AP*TP*AP*TP*AP*CP*TP*AP*AP*GP*AP*AP*C)-3')
| Macromolecule | Name: DNA (5'-D(P*TP*AP*TP*AP*TP*AP*CP*TP*AP*AP*GP*AP*AP*C)-3') type: dna / ID: 3 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: (DT)(DA)(DT)(DA)(DT)(DA)(DC)(DT)(DA)(DA) (DG)(DA)(DA)(DC) |
-Macromolecule #4: DNA (26-MER)
| Macromolecule | Name: DNA (26-MER) / type: dna / ID: 4 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: (DC)(DG)(DT)(DT)(DT)(DC)(DA)(DT)(DT)(DG) (DT)(DT)(DG)(DT)(DT)(DC)(DT)(DT)(DA)(DG) (DT)(DA)(DT)(DA)(DT)(DA) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 Component:
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R3.5/1 / Material: COPPER / Mesh: 200 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Support film - Film thickness: 200.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 101.325 kPa | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 11520 pixel / Digitization - Dimensions - Height: 8184 pixel / Number grids imaged: 2 / Number real images: 32723 / Average exposure time: 0.0426 sec. / Average electron dose: 46.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 30000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coefficient |
|---|
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)