+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23404 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Reconsituted 207 bp Archaeasome, Class II | |||||||||
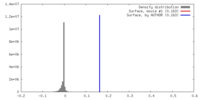
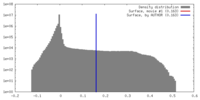
 Map data Map data | Reconstructed density of the Arc207 archaeasome complex. 7 histone dimers wrap 207 bp of DNA through a 4 dimer (Arc120) "base" and 3 dimer (Arc90) "lid" arrangement. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Thermococcus kodakarensis (archaea) Thermococcus kodakarensis (archaea) | |||||||||
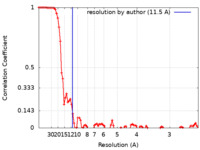
| Method | single particle reconstruction / cryo EM / Resolution: 11.5 Å | |||||||||
 Authors Authors | Bowerman S / Luger K | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: Archaeal chromatin 'slinkies' are inherently dynamic complexes with deflected DNA wrapping pathways. Authors: Samuel Bowerman / Jeff Wereszczynski / Karolin Luger /  Abstract: Eukaryotes and many archaea package their DNA with histones. While the four eukaryotic histones wrap ~147 DNA base pairs into nucleosomes, archaeal histones form 'nucleosome-like' complexes that ...Eukaryotes and many archaea package their DNA with histones. While the four eukaryotic histones wrap ~147 DNA base pairs into nucleosomes, archaeal histones form 'nucleosome-like' complexes that continuously wind between 60 and 500 base pairs of DNA ('archaeasomes'), suggested by crystal contacts and analysis of cellular chromatin. Solution structures of large archaeasomes (>90 DNA base pairs) have never been directly observed. Here, we utilize molecular dynamics simulations, analytical ultracentrifugation, and cryoEM to structurally characterize the solution state of archaeasomes on longer DNA. Simulations reveal dynamics of increased accessibility without disruption of DNA-binding or tetramerization interfaces. Mg concentration influences compaction, and cryoEM densities illustrate that DNA is wrapped in consecutive substates arranged 90 out-of-plane with one another. Without ATP-dependent remodelers, archaea may leverage these inherent dynamics to balance chromatin packing and accessibility. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23404.map.gz emd_23404.map.gz | 59.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23404-v30.xml emd-23404-v30.xml emd-23404.xml emd-23404.xml | 10.3 KB 10.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23404_fsc.xml emd_23404_fsc.xml | 9 KB | Display |  FSC data file FSC data file |
| Images |  emd_23404.png emd_23404.png | 58.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23404 http://ftp.pdbj.org/pub/emdb/structures/EMD-23404 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23404 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23404 | HTTPS FTP |
-Validation report
| Summary document |  emd_23404_validation.pdf.gz emd_23404_validation.pdf.gz | 321 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23404_full_validation.pdf.gz emd_23404_full_validation.pdf.gz | 320.6 KB | Display | |
| Data in XML |  emd_23404_validation.xml.gz emd_23404_validation.xml.gz | 11.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23404 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23404 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23404 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23404 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_23404.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23404.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstructed density of the Arc207 archaeasome complex. 7 histone dimers wrap 207 bp of DNA through a 4 dimer (Arc120) "base" and 3 dimer (Arc90) "lid" arrangement. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.219 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Archaeasome containing 207 bp of DNA
| Entire | Name: Archaeasome containing 207 bp of DNA |
|---|---|
| Components |
|
-Supramolecule #1: Archaeasome containing 207 bp of DNA
| Supramolecule | Name: Archaeasome containing 207 bp of DNA / type: complex / ID: 1 / Parent: 0 Details: Archaeasome (archaeal histone-DNA complex) formed by 7 histone dimers wrapping 207 bp of DNA. |
|---|---|
| Source (natural) | Organism:   Thermococcus kodakarensis (archaea) Thermococcus kodakarensis (archaea) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Quantifoil R2/2 / Pretreatment - Type: GLOW DISCHARGE / Details: 40 mA | |||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER Details: Manual plunge at ambient temperature and humidity.. | |||||||||
| Details | Sample was observed to be monodisperse via Sedimentation Velocity Analytical Ultracentrifugation. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 5388 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)