+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22365 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of GABAA receptor inside synapse | ||||||||||||
 Map data Map data | GABAA receptor inside synapse | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  | ||||||||||||
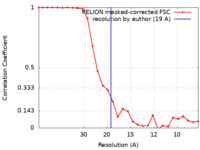
| Method | subtomogram averaging / cryo EM / Resolution: 19.0 Å | ||||||||||||
 Authors Authors | Liu YT / Tao CL / Zhang X / Xia W / Shi DQ / Qi L / Xu C / Sun R / Li XW / Lau PM ...Liu YT / Tao CL / Zhang X / Xia W / Shi DQ / Qi L / Xu C / Sun R / Li XW / Lau PM / Zhou ZH / Bi GQ | ||||||||||||
| Funding support |  China, China,  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Neurosci / Year: 2020 Journal: Nat Neurosci / Year: 2020Title: Mesophasic organization of GABA receptors in hippocampal inhibitory synapses. Authors: Yun-Tao Liu / Chang-Lu Tao / Xiaokang Zhang / Wenjun Xia / Dong-Qing Shi / Lei Qi / Cheng Xu / Rong Sun / Xiao-Wei Li / Pak-Ming Lau / Z Hong Zhou / Guo-Qiang Bi /   Abstract: Information processing in the brain depends on specialized organization of neurotransmitter receptors and scaffolding proteins within the postsynaptic density. However, how these molecules are ...Information processing in the brain depends on specialized organization of neurotransmitter receptors and scaffolding proteins within the postsynaptic density. However, how these molecules are organized in situ remains largely unknown. In this study, template-free classification of oversampled sub-tomograms was used to analyze cryo-electron tomograms of hippocampal synapses. We identified type-A GABA receptors (GABARs) in inhibitory synapses and determined their in situ structure at 19-Å resolution. These receptors are organized hierarchically: from GABAR super-complexes with a preferred inter-receptor distance of 11 nm but variable relative angles, through semi-ordered, two-dimensional receptor networks with reduced Voronoi entropy, to mesophasic assembly with a sharp phase boundary. These assemblies likely form via interactions among postsynaptic scaffolding proteins and receptors and align with putative presynaptic vesicle release sites. Such mesophasic self-organization might allow synapses to achieve a 'Goldilocks' state, striking a balance between stability and flexibility and enabling plasticity in information processing. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22365.map.gz emd_22365.map.gz | 926.4 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22365-v30.xml emd-22365-v30.xml emd-22365.xml emd-22365.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22365_fsc.xml emd_22365_fsc.xml | 2.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_22365.png emd_22365.png | 45.7 KB | ||
| Others |  emd_22365_half_map_1.map.gz emd_22365_half_map_1.map.gz emd_22365_half_map_2.map.gz emd_22365_half_map_2.map.gz | 720.4 KB 720.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22365 http://ftp.pdbj.org/pub/emdb/structures/EMD-22365 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22365 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22365 | HTTPS FTP |
-Validation report
| Summary document |  emd_22365_validation.pdf.gz emd_22365_validation.pdf.gz | 79.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22365_full_validation.pdf.gz emd_22365_full_validation.pdf.gz | 78.6 KB | Display | |
| Data in XML |  emd_22365_validation.xml.gz emd_22365_validation.xml.gz | 495 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22365 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22365 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22365 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22365 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22365.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22365.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GABAA receptor inside synapse | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: GABAA receptor inside synapse
| File | emd_22365_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GABAA receptor inside synapse | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: GABAA receptor inside synapse
| File | emd_22365_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GABAA receptor inside synapse | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : gamma-aminobutyric acid receptor
| Entire | Name: gamma-aminobutyric acid receptor |
|---|---|
| Components |
|
-Supramolecule #1: gamma-aminobutyric acid receptor
| Supramolecule | Name: gamma-aminobutyric acid receptor / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 250 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.3 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
| Details | This sample is cultured hippocampal neurons. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 2.5 e/Å2 Details: Tilt series were collected first from 0 to -60 and then from +2 to +60 at 2 intervals using FEI Xplore 3D software, with the defocus value set at -12 to -18 um, and the total electron dosage ...Details: Tilt series were collected first from 0 to -60 and then from +2 to +60 at 2 intervals using FEI Xplore 3D software, with the defocus value set at -12 to -18 um, and the total electron dosage of about 100 e-/A2. The final pixel size was 0.755 nm. For the analysis of GABAARs, cryoET data were collected using a Titan Krios (Thermo Fisher) equipped with a Volta phase plate (VPP), a post-column energy filter (Gatan image filter), and a K2 Summit direct electron detector (Gatan). The energy filter slit was set at 20 eV. The Titan Krios was operated at an acceleration voltage of 300 KV. When VPP was used, the defocus value was maintained at -1 um; otherwise, it was maintained at -4 um. The VPP was conditioned by pre-irradiation for 60 s to achieve an initial phase shift of about 0.3pi. Images were collected by the K2 camera in counting mode or super-resolution mode. When counting mode was used, the pixel size was 0.435 nm. For super-resolution mode image, the final pixel size was 0.265 nm. Tilt series were acquired using SerialEM49 with two tilt schemes: from +48 to -60 and from +50 to +66 at an interval of 2; from +48 to -60 and from +51 to +66 at an interval of 3. The total accumulated dose is ~150 e-/A2. For sub-tomogram analysis, 6 grids were used for data collection. Totally, 32 and 40 inhibitory synapses were imaged with and without VPP, respectively. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)