+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-22189 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | R. capsulatus cyt bc1 (CIII2) at 3.3A | ||||||||||||||||||
 マップデータ マップデータ | CIII2 | ||||||||||||||||||
 試料 試料 |
| ||||||||||||||||||
 キーワード キーワード | cytochrome bc1 membrane protein complex ubiquinone:cytochrome c oxidoreductase Complex III / OXIDOREDUCTASE | ||||||||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報respiratory chain complex III / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity / respiratory electron transport chain / 2 iron, 2 sulfur cluster binding / electron transfer activity / heme binding / metal ion binding / plasma membrane 類似検索 - 分子機能 | ||||||||||||||||||
| 生物種 |  Rhodobacter capsulatus SB 1003 (バクテリア) / Rhodobacter capsulatus SB 1003 (バクテリア) /  Rhodobacter capsulatus (strain ATCC BAA-309 / NBRC 16581 / SB1003) (バクテリア) Rhodobacter capsulatus (strain ATCC BAA-309 / NBRC 16581 / SB1003) (バクテリア) | ||||||||||||||||||
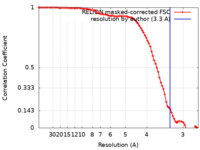
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.3 Å | ||||||||||||||||||
 データ登録者 データ登録者 | Steimle S / Van Eeuwen T | ||||||||||||||||||
| 資金援助 |  米国, 5件 米国, 5件
| ||||||||||||||||||
 引用 引用 |  ジャーナル: Nat Commun / 年: 2021 ジャーナル: Nat Commun / 年: 2021タイトル: Cryo-EM structures of engineered active bc-cbb type CIIICIV super-complexes and electronic communication between the complexes. 著者: Stefan Steimle / Trevor van Eeuwen / Yavuz Ozturk / Hee Jong Kim / Merav Braitbard / Nur Selamoglu / Benjamin A Garcia / Dina Schneidman-Duhovny / Kenji Murakami / Fevzi Daldal /    要旨: Respiratory electron transport complexes are organized as individual entities or combined as large supercomplexes (SC). Gram-negative bacteria deploy a mitochondrial-like cytochrome (cyt) bc (Complex ...Respiratory electron transport complexes are organized as individual entities or combined as large supercomplexes (SC). Gram-negative bacteria deploy a mitochondrial-like cytochrome (cyt) bc (Complex III, CIII), and may have specific cbb-type cyt c oxidases (Complex IV, CIV) instead of the canonical aa-type CIV. Electron transfer between these complexes is mediated by soluble (c) and membrane-anchored (c) cyts. Here, we report the structure of an engineered bc-cbb type SC (CIIICIV, 5.2 Å resolution) and three conformers of native CIII (3.3 Å resolution). The SC is active in vivo and in vitro, contains all catalytic subunits and cofactors, and two extra transmembrane helices attributed to cyt c and the assembly factor CcoH. The cyt c is integral to SC, its cyt domain is mobile and it conveys electrons to CIV differently than cyt c. The successful production of a native-like functional SC and determination of its structure illustrate the characteristics of membrane-confined and membrane-external respiratory electron transport pathways in Gram-negative bacteria. | ||||||||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_22189.map.gz emd_22189.map.gz | 4.3 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-22189-v30.xml emd-22189-v30.xml emd-22189.xml emd-22189.xml | 16.7 KB 16.7 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_22189_fsc.xml emd_22189_fsc.xml | 8.5 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_22189.png emd_22189.png | 244.3 KB | ||
| Filedesc metadata |  emd-22189.cif.gz emd-22189.cif.gz | 6.5 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22189 http://ftp.pdbj.org/pub/emdb/structures/EMD-22189 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22189 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22189 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_22189_validation.pdf.gz emd_22189_validation.pdf.gz | 391.4 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_22189_full_validation.pdf.gz emd_22189_full_validation.pdf.gz | 391 KB | 表示 | |
| XML形式データ |  emd_22189_validation.xml.gz emd_22189_validation.xml.gz | 10.3 KB | 表示 | |
| CIF形式データ |  emd_22189_validation.cif.gz emd_22189_validation.cif.gz | 13.6 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22189 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22189 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22189 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22189 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  6xi0MC  6xktC  6xkuC  6xkvC  6xkwC  6xkxC  6xkzC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | |
| 電子顕微鏡画像生データ |  EMPIAR-10735 (タイトル: Cryo-EM structures of engineered active bc1-cbb3 type CIII2CIV super-complexes and electronic communication between the complexes EMPIAR-10735 (タイトル: Cryo-EM structures of engineered active bc1-cbb3 type CIII2CIV super-complexes and electronic communication between the complexesData size: 35.8 TB Data #1: tripartite SC - dataset 1 [micrographs - multiframe] Data #2: tripartite SC - dataset 2 [micrographs - multiframe] Data #3: tripartite SC - dataset 3 [micrographs - multiframe] Data #4: tripartite SC - dataset 4 [micrographs - multiframe] Data #5: tripartite SC - dataset 5 [micrographs - multiframe] Data #6: tripartite SC - dataset 6 [micrographs - multiframe] Data #7: tripartite SC - dataset 7 [micrographs - multiframe] Data #8: bipartite SC - dataset 1 [micrographs - single frame] Data #9: bipartite SC - dataset 2 [micrographs - multiframe] Data #10: bipartite SC - dataset 3 [micrographs - multiframe]) |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_22189.map.gz / 形式: CCP4 / 大きさ: 52.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_22189.map.gz / 形式: CCP4 / 大きさ: 52.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | CIII2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : homodimer of ubiquinol:cytochrome c reductase (complex III or cyt...
| 全体 | 名称: homodimer of ubiquinol:cytochrome c reductase (complex III or cyt bc1 complex) |
|---|---|
| 要素 |
|
-超分子 #1: homodimer of ubiquinol:cytochrome c reductase (complex III or cyt...
| 超分子 | 名称: homodimer of ubiquinol:cytochrome c reductase (complex III or cyt bc1 complex) タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1-#3 |
|---|---|
| 由来(天然) | 生物種:  Rhodobacter capsulatus SB 1003 (バクテリア) Rhodobacter capsulatus SB 1003 (バクテリア) |
| 分子量 | 理論値: 250 KDa |
-分子 #1: Ubiquinol-cytochrome c reductase iron-sulfur subunit
| 分子 | 名称: Ubiquinol-cytochrome c reductase iron-sulfur subunit タイプ: protein_or_peptide / ID: 1 / コピー数: 2 / 光学異性体: LEVO / EC番号: quinol-cytochrome-c reductase |
|---|---|
| 由来(天然) | 生物種:  Rhodobacter capsulatus (strain ATCC BAA-309 / NBRC 16581 / SB1003) (バクテリア) Rhodobacter capsulatus (strain ATCC BAA-309 / NBRC 16581 / SB1003) (バクテリア)株: ATCC BAA-309 / NBRC 16581 / SB1003 |
| 分子量 | 理論値: 20.465109 KDa |
| 組換発現 | 生物種:  Rhodobacter capsulatus SB 1003 (バクテリア) Rhodobacter capsulatus SB 1003 (バクテリア) |
| 配列 | 文字列: MSHAEDNAGT RRDFLYHATA ATGVVVTGAA VWPLINQMNA SADVKAMASI FVDVSAVEVG TQLTVKWRGK PVFIRRRDEK DIELARSVP LGALRDTSAE NANKPGAEAT DENRTLPAFD GTNTGEWLVM LGVCTHLGCV PMGDKSGDFG GWFCPCHGSH Y DSAGRIRK ...文字列: MSHAEDNAGT RRDFLYHATA ATGVVVTGAA VWPLINQMNA SADVKAMASI FVDVSAVEVG TQLTVKWRGK PVFIRRRDEK DIELARSVP LGALRDTSAE NANKPGAEAT DENRTLPAFD GTNTGEWLVM LGVCTHLGCV PMGDKSGDFG GWFCPCHGSH Y DSAGRIRK GPAPRNLDIP VAAFVDETTI KLG UniProtKB: Ubiquinol-cytochrome c reductase iron-sulfur subunit |
-分子 #2: Cytochrome b
| 分子 | 名称: Cytochrome b / タイプ: protein_or_peptide / ID: 2 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Rhodobacter capsulatus (strain ATCC BAA-309 / NBRC 16581 / SB1003) (バクテリア) Rhodobacter capsulatus (strain ATCC BAA-309 / NBRC 16581 / SB1003) (バクテリア)株: ATCC BAA-309 / NBRC 16581 / SB1003 |
| 分子量 | 理論値: 49.386469 KDa |
| 組換発現 | 生物種:  Rhodobacter capsulatus SB 1003 (バクテリア) Rhodobacter capsulatus SB 1003 (バクテリア) |
| 配列 | 文字列: MSGIPHDHYE PKTGIEKWLH DRLPIVGLVY DTIMIPTPKN LNWWWIWGIV LAFTLVLQIV TGIVLAMHYT PHVDLAFASV EHIMRDVNG GWAMRYIHAN GASLFFLAVY IHIFRGLYYG SYKAPREITW IVGMVIYLLM MGTAFMGYVL PWGQMSFWGA T VITGLFGA ...文字列: MSGIPHDHYE PKTGIEKWLH DRLPIVGLVY DTIMIPTPKN LNWWWIWGIV LAFTLVLQIV TGIVLAMHYT PHVDLAFASV EHIMRDVNG GWAMRYIHAN GASLFFLAVY IHIFRGLYYG SYKAPREITW IVGMVIYLLM MGTAFMGYVL PWGQMSFWGA T VITGLFGA IPGIGPSIQA WLLGGPAVDN ATLNRFFSLH YLLPFVIAAL VAIHIWAFHT TGNNNPTGVE VRRTSKADAE KD TLPFWPY FVIKDLFALA LVLLGFFAVV AYMPNYLGHP DNYVQANPLS TPAHIVPEWY FLPFYAILRA FAADVWVVIL VDG LTFGIV DAKFFGVIAM FGAIAVMALA PWLDTSKVRS GAYRPKFRMW FWFLVLDFVV LTWVGAMPTE YPYDWISLIA STYW FAYFL VILPLLGATE KPEPIPASIE EDFNSHYGNP AE UniProtKB: Cytochrome b |
-分子 #3: Cytochrome c1
| 分子 | 名称: Cytochrome c1 / タイプ: protein_or_peptide / ID: 3 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Rhodobacter capsulatus (strain ATCC BAA-309 / NBRC 16581 / SB1003) (バクテリア) Rhodobacter capsulatus (strain ATCC BAA-309 / NBRC 16581 / SB1003) (バクテリア)株: ATCC BAA-309 / NBRC 16581 / SB1003 |
| 分子量 | 理論値: 30.352615 KDa |
| 組換発現 | 生物種:  Rhodobacter capsulatus SB 1003 (バクテリア) Rhodobacter capsulatus SB 1003 (バクテリア) |
| 配列 | 文字列: MKKLLISAVS ALVLGSGAAF ANSNVPDHAF SFEGIFGKYD QAQLRRGFQV YNEVCSACHG MKFVPIRTLA DDGGPQLDPT FVREYAAGL DTIIDKDSGE ERDRKETDMF PTRVGDGMGP DLSVMAKARA GFSGPAGSGM NQLFKGMGGP EYIYNYVIGF E ENPECAPE ...文字列: MKKLLISAVS ALVLGSGAAF ANSNVPDHAF SFEGIFGKYD QAQLRRGFQV YNEVCSACHG MKFVPIRTLA DDGGPQLDPT FVREYAAGL DTIIDKDSGE ERDRKETDMF PTRVGDGMGP DLSVMAKARA GFSGPAGSGM NQLFKGMGGP EYIYNYVIGF E ENPECAPE GIDGYYYNKT FQIGGVPDTC KDAAGVKITH GSWARMPPPL VDDQVTYEDG TPATVDQMAQ DVSAFLMWAA EP KLVARKQ MGLVAMVMLG LLSVMLYLTN KRLWAPYKGH KA UniProtKB: Cytochrome c1 |
-分子 #4: FE2/S2 (INORGANIC) CLUSTER
| 分子 | 名称: FE2/S2 (INORGANIC) CLUSTER / タイプ: ligand / ID: 4 / コピー数: 2 / 式: FES |
|---|---|
| 分子量 | 理論値: 175.82 Da |
| Chemical component information |  ChemComp-FES: |
-分子 #5: HEME C
| 分子 | 名称: HEME C / タイプ: ligand / ID: 5 / コピー数: 6 / 式: HEC |
|---|---|
| 分子量 | 理論値: 618.503 Da |
| Chemical component information |  ChemComp-HEC: |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 4 mg/mL |
|---|---|
| 緩衝液 | pH: 7.4 |
| グリッド | モデル: C-flat-1.2/1.3 / 材質: COPPER / メッシュ: 400 / 前処理 - タイプ: GLOW DISCHARGE |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 298 K / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 (6k x 4k) / 平均電子線量: 40.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: SPOT SCAN / 撮影モード: BRIGHT FIELD |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)